| Sequence ID | dm3.chr3R |
|---|---|
| Location | 2,056,322 – 2,056,393 |
| Length | 71 |
| Max. P | 0.663051 |

| Location | 2,056,322 – 2,056,393 |
|---|---|
| Length | 71 |
| Sequences | 11 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Shannon entropy | 0.39423 |
| G+C content | 0.28495 |
| Mean single sequence MFE | -12.81 |
| Consensus MFE | -7.90 |
| Energy contribution | -8.51 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 2056322 71 + 27905053 AUUAAUUCAAA-UGGCCGUAAAA-----GGGAAAAUGU-GUUCUA------UUAACUUGGCAUUUUUAUGGCUUUGUGAAAUCU .....((((..-.((((((((((-----(.(..((.((-......------...))))..).)))))))))))...)))).... ( -16.20, z-score = -2.46, R) >droSim1.chr3R 2072852 71 + 27517382 AUUAAUUCAAA-UGGCCGUAAAA-----GGGAAAAUGU-GUUCUA------UUAACUUGACAUUUUUAUGGCUUUGUGAAAUCU .....((((..-.((((((((((-----(.......((-......------...))......)))))))))))...)))).... ( -15.62, z-score = -2.55, R) >droSec1.super_6 2142891 71 + 4358794 AUUAAUUCAAA-UGGCCGUAAAA-----GGGAAAAUGU-GUUCUA------UUAACUUGACAUUUUUAUGGCUUUGUGAAAUCU .....((((..-.((((((((((-----(.......((-......------...))......)))))))))))...)))).... ( -15.62, z-score = -2.55, R) >droEre2.scaffold_4770 2298180 71 + 17746568 AUUAAUUCAAA-UGGCCGUAAAA-----GGGAAAAUGU-GUUCUA------UUAACUUGACAUUUUUAUGGCUUUGUGAAAUCU .....((((..-.((((((((((-----(.......((-......------...))......)))))))))))...)))).... ( -15.62, z-score = -2.55, R) >droYak2.chr3R 17982759 62 + 28832112 AUUAAUUCAAA-UGGCUGAAAAA-----GGGAAAAUGU-AUUCUA------UUAACUUGACAUUUUUAUGGCUUU--------- .....((((..-....))))(((-----(.((((((((-......------........))))))))....))))--------- ( -5.04, z-score = 0.62, R) >droAna3.scaffold_13340 5595352 71 - 23697760 AUUAAUUCAAA-UGGCCGUAAAA-----GGGAAAAUGU-GUUCUA------UUAACUUGACAUUUUUAUGGGUUUGUGAAAUCU .....((((..-.(.((((((((-----(.......((-......------...))......))))))))).)...)))).... ( -11.12, z-score = -0.47, R) >dp4.chr2 15021511 71 - 30794189 AUUAAUUCAAA-UGACCGUAAAA-----GGGAAAAUCU-GUUCUA------UUAACUUGACAUUUUUAUGGUUUUGUGAAAUCU .....((((..-.((((((((((-----(((((.....-.)))).------...........)))))))))))...)))).... ( -12.80, z-score = -1.52, R) >droPer1.super_0 4641205 69 + 11822988 AUUAAUUCAA--UGACCGUAAAA-----GGGAAA-UCU-GUUCUA------UUAACUUGACAUUUUUAUGGUUUUGUGAAAUCU .....((((.--.((((((((((-----(.....-...-(((...------..)))......)))))))))))...)))).... ( -13.04, z-score = -1.66, R) >droWil1.scaffold_181130 5403057 83 - 16660200 AUUAAUUCAAUGUGACCGUAAAAAAAAGGGGAAAAUAGAGUUCUACGAGUAUUAGCUUAACAUUUUUAUGGUUU-GUGAAAUUU .....((((....(((((((((((...(..(((.......)))..)((((....))))....))))))))))).-.)))).... ( -16.40, z-score = -2.01, R) >droVir3.scaffold_12822 2276000 71 - 4096053 UACAAUUCAAA-UGAACAUAAAA------GGCGAAUGAGAUUAAA-----GUUUAUUAACCAAGUUUAUGGCUU-GUGAAAUUU .....((((..-....((((((.------((..((((((......-----.))))))..))...))))))....-.)))).... ( -10.20, z-score = -1.01, R) >droMoj3.scaffold_6540 11322802 71 + 34148556 UACAAUUCAAA-UAAACGUAAAA------GGCGAAUGAGAUUAAA-----GUUUACCAACCAAGUUUAUGGCUU-GUGAAAUUU (((((..((..-(((((......------((..(((.........-----)))..))......)))))))..))-)))...... ( -9.30, z-score = -0.46, R) >consensus AUUAAUUCAAA_UGGCCGUAAAA_____GGGAAAAUGU_GUUCUA______UUAACUUGACAUUUUUAUGGCUUUGUGAAAUCU .....((((....((((((((((.....(((........................))).....))))))))))...)))).... ( -7.90 = -8.51 + 0.62)


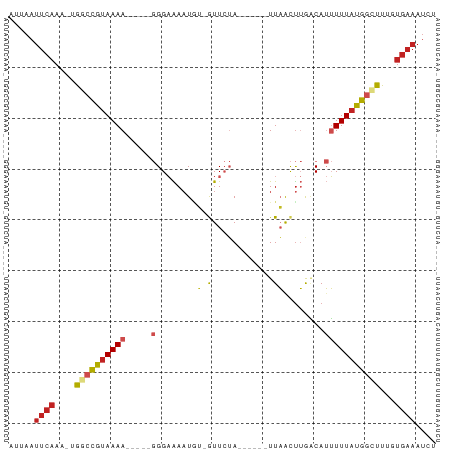
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:10 2011