
| Sequence ID | dm3.chr2L |
|---|---|
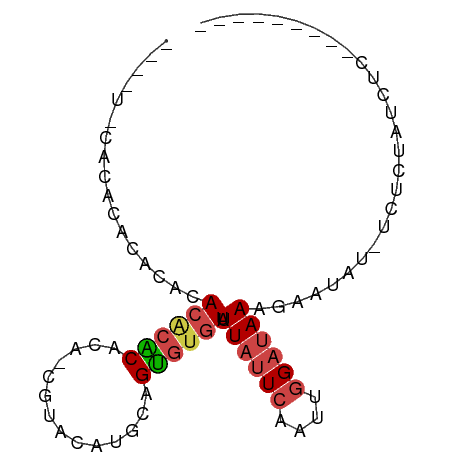
| Location | 6,455,160 – 6,455,237 |
| Length | 77 |
| Max. P | 0.998059 |

| Location | 6,455,160 – 6,455,237 |
|---|---|
| Length | 77 |
| Sequences | 6 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 68.08 |
| Shannon entropy | 0.55622 |
| G+C content | 0.36599 |
| Mean single sequence MFE | -13.37 |
| Consensus MFE | -5.92 |
| Energy contribution | -6.39 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
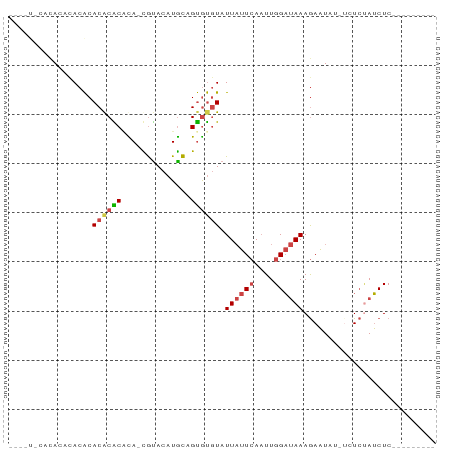
>dm3.chr2L 6455160 77 + 23011544 UAUUUACACACGCACACACACGCACA-CGUACAUGCAGUGUGUAUUAUUCAAUUGGAUAAAGAAUCUAUCUCUAUCUC--------- ....((((((((((.....(((....-)))...))).)))))))..........(((((.(((.....))).))))).--------- ( -16.10, z-score = -2.79, R) >droYak2.chr2L 15861946 72 - 22324452 --CAUUCACAAACACACACACACGUA-UGUACAUACAGUGUGUAUUAUUCAAUUGGAUAAAGAAUCG---UCUAUCUC--------- --.((((..........(((((((((-(....)))).)))))).((((((....)))))).))))..---........--------- ( -14.60, z-score = -1.97, R) >droEre2.scaffold_4929 15363549 73 + 26641161 --CAUUCAUACCCACACACACACUCACUGUACAUGCAGUGUGUAUUAUUCAAUUGGAUAAAGAAUAG---UCCAUCUC--------- --.((((..............((.((((((....)))))).)).((((((....)))))).))))..---........--------- ( -14.00, z-score = -1.48, R) >droSec1.super_5 4504341 69 + 5866729 --------UAUUUACACACGCACACA-CGUACAUGCCGUGUGUAUUAUUCAAUUGGAUAAAGAAUUUAUCUCUAUCUC--------- --------....((((((((((....-......)).))))))))..........(((((.(((.....))).))))).--------- ( -14.30, z-score = -2.66, R) >droSim1.chr2L 6241174 69 + 22036055 --------UAUUUACACACGCACACA-CGUACAUGCCGUGUGUAUUAUUCAAUUGGAUAAAGAAUUUAUCUCUAUCUC--------- --------....((((((((((....-......)).))))))))..........(((((.(((.....))).))))).--------- ( -14.30, z-score = -2.66, R) >droMoj3.scaffold_6540 14898127 84 + 34148556 -UAGCACACACACACGCACACGCAUA-CACAAAUGUAGCACAUAUUUAUAACUUGGCCAAAAAUUAU-UGUAGAGCUCAGUCACUUC -..((..........((....)).((-((....))))))...........(((.((((((......)-))....))).)))...... ( -6.90, z-score = 1.43, R) >consensus ____U_CACACACACACACACACACA_CGUACAUGCAGUGUGUAUUAUUCAAUUGGAUAAAGAAUAU_UCUCUAUCUC_________ .................((((((..............)))))).((((((....))))))........................... ( -5.92 = -6.39 + 0.47)

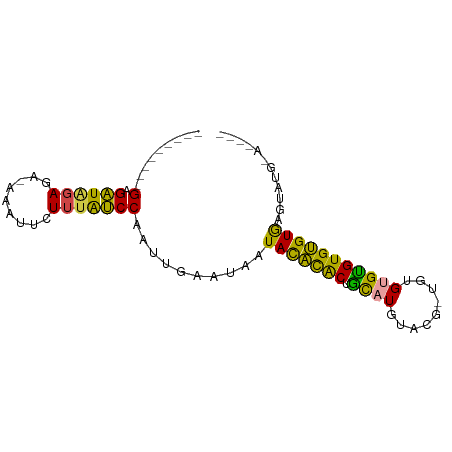
| Location | 6,455,160 – 6,455,237 |
|---|---|
| Length | 77 |
| Sequences | 6 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 68.08 |
| Shannon entropy | 0.55622 |
| G+C content | 0.36599 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -12.05 |
| Energy contribution | -10.67 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
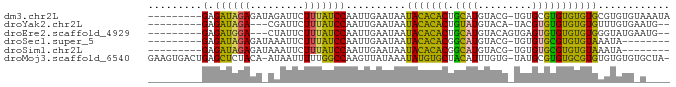
>dm3.chr2L 6455160 77 - 23011544 ---------GAGAUAGAGAUAGAUUCUUUAUCCAAUUGAAUAAUACACACUGCAUGUACG-UGUGCGUGUGUGUGCGUGUGUAAAUA ---------(.((((((((.....)))))))))..........(((((((((((..((((-....))))..)))).))))))).... ( -27.50, z-score = -4.32, R) >droYak2.chr2L 15861946 72 + 22324452 ---------GAGAUAGA---CGAUUCUUUAUCCAAUUGAAUAAUACACACUGUAUGUACA-UACGUGUGUGUGUGUUUGUGAAUG-- ---------...(((((---(.((((...........)))).(((((((((((((....)-)))).)))))))))))))).....-- ( -18.20, z-score = -1.60, R) >droEre2.scaffold_4929 15363549 73 - 26641161 ---------GAGAUGGA---CUAUUCUUUAUCCAAUUGAAUAAUACACACUGCAUGUACAGUGAGUGUGUGUGUGGGUAUGAAUG-- ---------........---.(((((..((((((........(((((((((.(((.....)))))))))))).)))))).)))))-- ( -20.60, z-score = -1.91, R) >droSec1.super_5 4504341 69 - 5866729 ---------GAGAUAGAGAUAAAUUCUUUAUCCAAUUGAAUAAUACACACGGCAUGUACG-UGUGUGCGUGUGUAAAUA-------- ---------(.((((((((.....)))))))))..........((((((((((((.....-.)))).))))))))....-------- ( -22.00, z-score = -4.05, R) >droSim1.chr2L 6241174 69 - 22036055 ---------GAGAUAGAGAUAAAUUCUUUAUCCAAUUGAAUAAUACACACGGCAUGUACG-UGUGUGCGUGUGUAAAUA-------- ---------(.((((((((.....)))))))))..........((((((((((((.....-.)))).))))))))....-------- ( -22.00, z-score = -4.05, R) >droMoj3.scaffold_6540 14898127 84 - 34148556 GAAGUGACUGAGCUCUACA-AUAAUUUUUGGCCAAGUUAUAAAUAUGUGCUACAUUUGUG-UAUGCGUGUGCGUGUGUGUGUGCUA- ...((((((..(((.....-.........)))..))))))......(..((((((.((..-((....))..)).))))).)..)..- ( -16.54, z-score = 1.05, R) >consensus _________GAGAUAGAGA_AAAUUCUUUAUCCAAUUGAAUAAUACACACUGCAUGUACG_UGUGUGUGUGUGUGAGUAUG_A____ .........(.((((((.........)))))))..........(((((((.(((((((...))))))))))))))............ (-12.05 = -10.67 + -1.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:20 2011