| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,961,077 – 1,961,175 |
| Length | 98 |
| Max. P | 0.820368 |

| Location | 1,961,077 – 1,961,175 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.66088 |
| G+C content | 0.59001 |
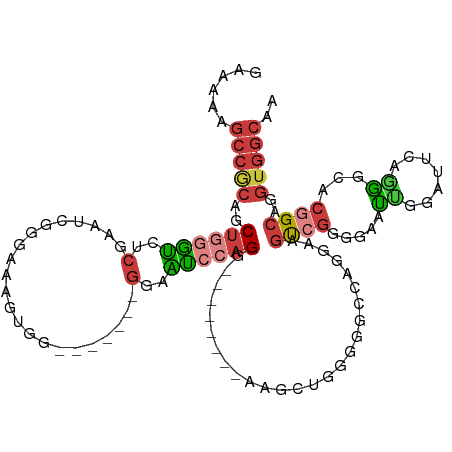
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -11.15 |
| Energy contribution | -12.32 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
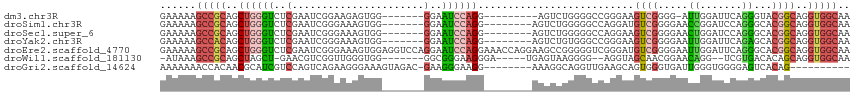
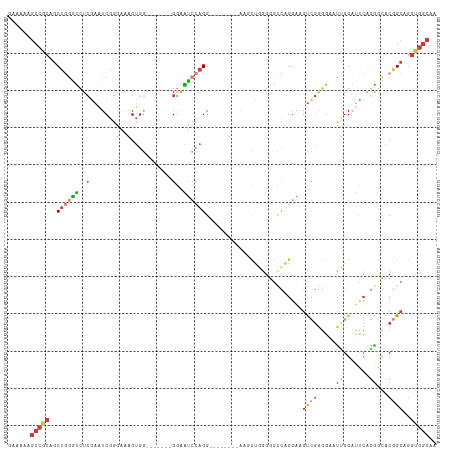
>dm3.chr3R 1961077 98 - 27905053 GAAAAAGCCGCAGCUGGGUCUCGAAUCGGAAGAGUGG-------GGAAUCCAGG---------AGUCUGGGGCCGGGAAGUCGGGG-AUUGGAUUCAGGGUACGGCAGGUGGCAA ......(((((.((((..((((.(.((....)).).)-------)))((((..(---------((((..(..((((....))))..-.)..))))).)))).))))..))))).. ( -38.20, z-score = -3.15, R) >droSim1.chr3R 1988314 100 - 27517382 GAAAAAGCCGCAGCUGGGUCUCGAAUCGGGAAAGUGG-------GGAAUCCAGG--------AGUCUGGGGGCCAGGAUGUCGGGGAACCGGAUCCAGGGCACGGCAGGUGGCAA ......(((((.((((.((((................-------((..(((((.--------...)))))..)).((((.((((....)))))))).)))).))))..))))).. ( -40.60, z-score = -2.07, R) >droSec1.super_6 2061869 100 - 4358794 GAAAAAGCCGCAGCUGGGUCUCGAAUCGGGAAAGUGG-------GGAAUCCAGG--------AGUCUGGGGGCCAGGAAGUCGGGGAACUGGAUCCAGGGCACGGCAGGUGGCAA ......(((((.((((.((((................-------((..(((((.--------...)))))..)).(((..((((....)))).))).)))).))))..))))).. ( -36.70, z-score = -1.39, R) >droYak2.chr3R 17904374 100 - 28832112 GAAAAAGCCACAGCUGGGUCUCGAAUCGGGAAAGUGG-------GGAAUCCAGG--------AGUCUGUGGGCCGGGAAGUCGGGGAAUUGGAUUCAGAGCACGGCAGGUGGCAA ......(((((.((((..(((((...)))))..((..-------.((((((((.--------..(((.(((.(......)))).))).))))))))...)).))))..))))).. ( -35.50, z-score = -2.26, R) >droEre2.scaffold_4770 2220344 115 - 17746568 GAAAAAGCCGCAGCUGGGUCUCGAAUCGGGAAAGUGGAGGUCCAGGAAUCCAGGAAACCAGGAAGCCGGGGGUCGGGAUGUCGGGGAAUUGGAUUCAGGGCACGGCAGGUGGCAA ......(((((.((((..(((((...)))))........((((..((((((((....((..((..((.(....).))...)).))...)))))))).)))).))))..))))).. ( -41.60, z-score = -1.87, R) >droWil1.scaffold_181130 5324855 97 + 16660200 -AUAAAGCCGCAGCUAGCU-GAACGUCGGUUGGGUGG-------GGCGGGAAGGGA-----UGAGUAAGGGG--AGGUAGCAACGGAACAGG--UCGUGACACAGCAGGUGGCAA -.....(((.((.((((((-(.....))))))).)).-------))).........-----...........--.....((.((((......--))))..(((.....))))).. ( -25.40, z-score = -0.90, R) >droGri2.scaffold_14624 1326780 96 + 4233967 AAAAAAACCACAACGCAUCGUCCAGUCAGAAGGGAAAGUAGAC-GAAGGGAAGG--------AAAGGCAGGUUGAAGCAGUGGGUGAUUGGGUGGGGAGUCACAG---------- .......((((..(.(.(((((...((......)).....)))-)).).)....--------....((........)).))))((((((........))))))..---------- ( -20.30, z-score = -1.01, R) >consensus GAAAAAGCCGCAGCUGGGUCUCGAAUCGGGAAAGUGG_______GGAAUCCAGG________AAGCUGGGGGCCAGGAAGUCGGGGAAUUGGAUUCAGGGCACGGCAGGUGGCAA ......(((((..((((((..(......................)..))))))..........................((((...................))))..))))).. (-11.15 = -12.32 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:02 2011