| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,954,106 – 1,954,207 |
| Length | 101 |
| Max. P | 0.804872 |

| Location | 1,954,106 – 1,954,207 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.50824 |
| G+C content | 0.43962 |
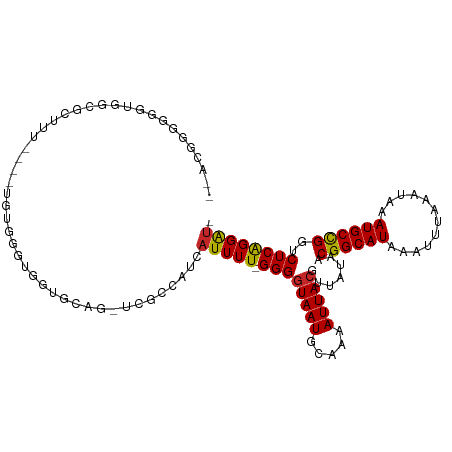
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.28 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.13 |
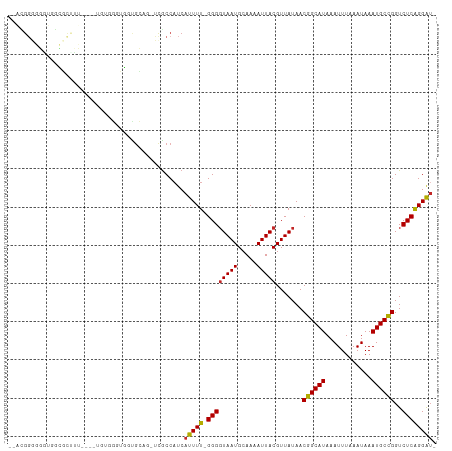
| Mean z-score | -2.01 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
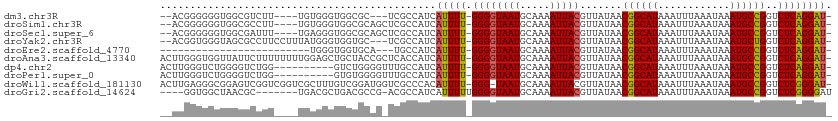
>dm3.chr3R 1954106 101 + 27905053 --ACGGGGGGUGGCGUCUU----UGUGGGUGGCGC---UCGCCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU- --..((.(((((.(..((.----...))..).)))---)).))...(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -33.00, z-score = -2.81, R) >droSim1.chr3R 1981405 104 + 27517382 --ACGGGGGGUGGCGCCUU----UGUGGGUGGCGCAGCUCGCCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU- --..((.((((.(((((..----.......))))).)))).))...(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -36.80, z-score = -2.83, R) >droSec1.super_6 2055019 104 + 4358794 --ACGGGGGGUGGCGAUUU----UGAGGGUGGCGCAGCUCGCCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU- --......((((((((..(----((.(.....).))).))))))))(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -29.80, z-score = -1.76, R) >droYak2.chr3R 17897149 105 + 28832112 --ACGGUGGGUAGCGCCUUCCUUUAUGGGUGGUGC---UCGCCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCUGGUCUCAGGAU- --..((((((((.(((((........))))).)))---)))))...(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -36.30, z-score = -3.53, R) >droEre2.scaffold_4770 2213550 82 + 17746568 -------------------------UGGGUGGUGCA---UGCCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU- -------------------------..((((((...---.))))))(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -24.30, z-score = -2.61, R) >droAna3.scaffold_13340 5514632 110 - 23697760 ACUUGGGUGGUUAUUCUUUUUUUUGGAGCUGCUACCGCUCACCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU- ...((((((((....((((.....)))).....)))))))).....(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -28.20, z-score = -1.85, R) >dp4.chr2 14940426 100 - 30794189 ACUUGGGUCUGGGGUCUGG----------GUCUGGGGUUUGCCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU- .((..(..(..(...)..)----------..)..))..........(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -26.70, z-score = -1.52, R) >droPer1.super_0 4558895 100 + 11822988 ACUUGGGUCUGGGGUCUGG----------GUGUGGGGUUUGCCAUCAUUUU-GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU- (((..(..(...)..)..)----------))((((......)))).(((((-((((((((.....))))).......((((((............))))))..))))))))- ( -25.70, z-score = -1.15, R) >droWil1.scaffold_181130 5315324 109 - 16660200 ACUUGAGGGCGGAGUCGGUCGGUCGCUUUGUCGGAUGGUCGCCCACAUUUU-GGG-UAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCGGGAU- .(((((((((((.(((.(.(((.....))).).)))..)))))).......-..(-((((.....))))).......((((((............))))))...)))))..- ( -31.50, z-score = -0.96, R) >droGri2.scaffold_14624 1319897 100 - 4233967 ----GGUGGCUAACGC-------UGACGCUGACGCCG-ACGCCAUCAUUUUUGGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCGGGGAU ----(..(((....))-------)..).....(.(((-(.(((.........(..((((((........))))))..)(((((............)))))))).)))).).. ( -27.70, z-score = -1.05, R) >consensus __ACGGGGGGUGGCGCUUU____UGUGGGUGGUGCAG_UCGCCAUCAUUUU_GGGGUAAUGCAAAAUUACGUUAUAACGGCAUAAAUUUAAAUAAAUGCCGGUCUCAGGAU_ ........................................(((.........(..((((((........))))))..)(((((............))))))))......... (-12.36 = -12.28 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:53:01 2011