| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,950,201 – 1,950,306 |
| Length | 105 |
| Max. P | 0.727079 |

| Location | 1,950,201 – 1,950,306 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.09 |
| Shannon entropy | 0.47352 |
| G+C content | 0.39125 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.89 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
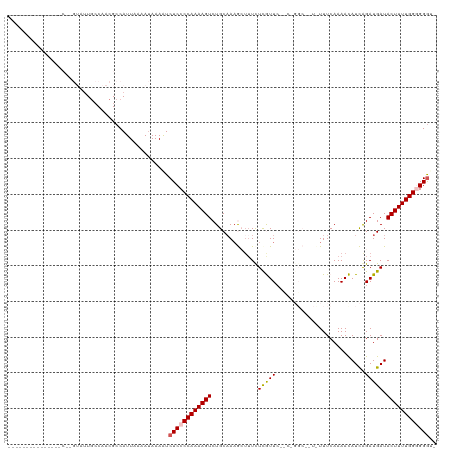
>dm3.chr3R 1950201 105 + 27905053 --------------GCAUGUAUUGCCAACGCCAUUAAAAUAAAGUCUCCCCAAAAGUCUGCAAAGCUUUCUCGUUCGCCCGGCCCUCUUUCAAAACAUCAGAUGAUACUUUUGGGGGAU- --------------(((.....)))..................(((.((((((((((..((...((..........))...)).............(((....))))))))))))))))- ( -20.90, z-score = -0.20, R) >droEre2.scaffold_4770 2209219 120 + 17746568 AUAAUGAACUGGUCUCUAGUAUUGCCAACGCCAUUCAAAUAAAAUCUCCCCAAAAGUCUGCAAAGCUUUUUUAUUAGCCCGGCCCUCUUUCAAAAUAUCAGAUGAUACUUUUGGAGGGGU ....((((.((((................))))))))........(((((((((((...((...(((........)))...))............((((....))))))))))).)))). ( -23.39, z-score = -0.05, R) >droYak2.chr3R 17893302 120 + 28832112 AUAAUAAACUGGUCUCCAGUUUUACCAACGCCAUUCAAAUAAAAUCUCCCCAAAAGUCUGCAAUGCGUUUUCAUUCUUCUGGCUUUUUUUCAAAAUAUCAGAUGAUACUUUUGGAGGGGU .....(((((((...))))))).........................(((((((((((.(.((((......)))))....))))))).(((((((((((....)))).))))))))))). ( -24.50, z-score = -1.46, R) >droSec1.super_6 2051102 87 + 4358794 ---------------------AUGGCAACGCCAUUAAAAUAAAGUCUCCCCAAAAGUCUGCUAAAAUUUCUCGUUC-----------UUUCAAAACAUUGGAUGAUACUUUUGGGGGGU- ---------------------(((((...)))))...........((((((((((((.............((((((-----------............)))))).)))))))))))).- ( -23.14, z-score = -2.59, R) >droSim1.chr3R 1977483 87 + 27517382 --------------------AUUGCCAACGCCAUUAAAA-AAAAUCUCCCCAAAAGUCUGCAAAAAUGUCUCGUUC-----------UUUCAAAACAUUGGAUGAUACUUUUGGGGGGU- --------------------...................-.....((((((((((((..(((....))).((((((-----------............)))))).)))))))))))).- ( -21.10, z-score = -2.00, R) >consensus _______________C__GUAUUGCCAACGCCAUUAAAAUAAAAUCUCCCCAAAAGUCUGCAAAGCUUUCUCGUUC__C_GGC__U_UUUCAAAACAUCAGAUGAUACUUUUGGGGGGU_ .............................................((((((((((((.............(((((.........................))))).)))))))))))).. (-12.69 = -12.89 + 0.20)


| Location | 1,950,201 – 1,950,306 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.09 |
| Shannon entropy | 0.47352 |
| G+C content | 0.39125 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -14.44 |
| Energy contribution | -13.56 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
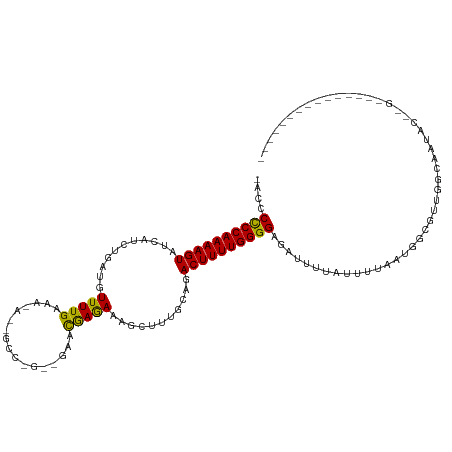
>dm3.chr3R 1950201 105 - 27905053 -AUCCCCCAAAAGUAUCAUCUGAUGUUUUGAAAGAGGGCCGGGCGAACGAGAAAGCUUUGCAGACUUUUGGGGAGACUUUAUUUUAAUGGCGUUGGCAAUACAUGC-------------- -.(((((((((((((((....))).............((..(((..........)))..))..)))))))))).)).............((((.(......)))))-------------- ( -24.80, z-score = 0.28, R) >droEre2.scaffold_4770 2209219 120 - 17746568 ACCCCUCCAAAAGUAUCAUCUGAUAUUUUGAAAGAGGGCCGGGCUAAUAAAAAAGCUUUGCAGACUUUUGGGGAGAUUUUAUUUGAAUGGCGUUGGCAAUACUAGAGACCAGUUCAUUAU .((((....((((((((....))))))))..(((((.((..((((........))))..))...)))))))))..........((((((((.((((.....)))).).))).)))).... ( -30.00, z-score = -0.80, R) >droYak2.chr3R 17893302 120 - 28832112 ACCCCUCCAAAAGUAUCAUCUGAUAUUUUGAAAAAAAGCCAGAAGAAUGAAAACGCAUUGCAGACUUUUGGGGAGAUUUUAUUUGAAUGGCGUUGGUAAAACUGGAGACCAGUUUAUUAU (((.((((((((((.((.((((...((((....))))..)))).))........((...))..))))))))))..((((.....))))......))).(((((((...)))))))..... ( -27.00, z-score = -1.23, R) >droSec1.super_6 2051102 87 - 4358794 -ACCCCCCAAAAGUAUCAUCCAAUGUUUUGAAA-----------GAACGAGAAAUUUUAGCAGACUUUUGGGGAGACUUUAUUUUAAUGGCGUUGCCAU--------------------- -...((((((((((...........(((((...-----------...)))))...........)))))))))).............(((((...)))))--------------------- ( -22.15, z-score = -2.40, R) >droSim1.chr3R 1977483 87 - 27517382 -ACCCCCCAAAAGUAUCAUCCAAUGUUUUGAAA-----------GAACGAGACAUUUUUGCAGACUUUUGGGGAGAUUUU-UUUUAAUGGCGUUGGCAAU-------------------- -...((((((((((..((...(((((((((...-----------...)))))))))..))...)))))))))).......-........((....))...-------------------- ( -25.30, z-score = -2.83, R) >consensus _ACCCCCCAAAAGUAUCAUCUGAUGUUUUGAAA_A__GCC_G__GAACGAGAAAGCUUUGCAGACUUUUGGGGAGAUUUUAUUUUAAUGGCGUUGGCAAUAC__G_______________ ....((((((((((...........(((((.................)))))...........))))))))))............................................... (-14.44 = -13.56 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:59 2011