
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,941,789 – 1,941,884 |
| Length | 95 |
| Max. P | 0.758399 |

| Location | 1,941,789 – 1,941,884 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Shannon entropy | 0.40809 |
| G+C content | 0.51793 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -21.88 |
| Energy contribution | -21.99 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
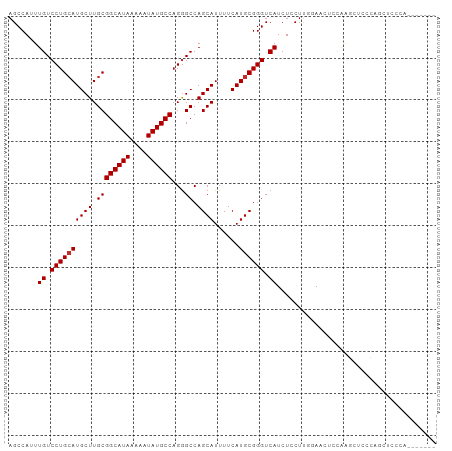
>dm3.chr3R 1941789 95 + 27905053 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUCGAACUCCGAGCUCCCAGCUCCAA------- .......((.((((((((((.((((((((....))))))...)).))))......)))))).))...............((((.....))))...------- ( -27.70, z-score = -0.97, R) >droSim1.chr3R 1969375 102 + 27517382 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUGGAACUCCGAGCUCCCAGCUCCCAGCUCCAA .......((.((((((((((.((((((((....))))))...)).))))......)))))).))......((((.((..((((.....))))..)).)))). ( -33.70, z-score = -1.51, R) >droSec1.super_6 2043082 102 + 4358794 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUGGAACUCCGAGCUCCCAGCUCCCAGCUCCAA .......((.((((((((((.((((((((....))))))...)).))))......)))))).))......((((.((..((((.....))))..)).)))). ( -33.70, z-score = -1.51, R) >droYak2.chr3R 17885077 102 + 28832112 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUGGAACUCCGAGCUCCCAGCUCCCAGCUCCAA .......((.((((((((((.((((((((....))))))...)).))))......)))))).))......((((.((..((((.....))))..)).)))). ( -33.70, z-score = -1.51, R) >droAna3.scaffold_13340 5503461 86 - 23697760 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUGGAGCCGUUGCUUCA---------------- .......((.((((((((((.((((((((....))))))...)).))))......)))))).))......((((((....))))))---------------- ( -27.90, z-score = -0.61, R) >dp4.chr2 14929321 92 - 30794189 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUACUUGCAGCUCCAGUCCCAGUCCCA---------- .(..(((.(..(((...(((.((((((((....))))))..((((.((((....)))).))))........)))))..)))..).)))..).---------- ( -26.40, z-score = -0.44, R) >droWil1.scaffold_181130 5301908 92 - 16660200 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGAGCCAGCAUUUUCAUGCGGGUCAUCUACUUGCAUCUCCAACUCUGGCUACG---------- (((((.......(((..((((..((((((....)))))).))))..))).....((((((((.....)))))))).........)))))...---------- ( -28.90, z-score = -2.26, R) >droMoj3.scaffold_6540 11216486 99 + 34148556 AGCCAUUUGUCCUGGAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUACUCCCAGCUACAGUUCCAGCUACAACUAGCA--- .((...((((.(((((.(((.((((((((....))))))..((((.((((....)))).))))...........))...))))))))..))))...)).--- ( -31.70, z-score = -1.70, R) >droGri2.scaffold_14624 1306050 96 - 4233967 AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUACUCCCAGCUAUACAGUUGCCCAGUUUCC------ .......((.((((((((((.((((((((....))))))...)).))))......)))))).))...(((..(((((....)))))...)))....------ ( -25.70, z-score = -0.31, R) >consensus AGCCAUUUGUCCUGCAUGCUUGCGGCAUAAAAAUAUGCCAGGGCCAGCAUUUUCAUGCGGGUCAUCUCCUUGGAACUCCAAGCUCCCAGCUCCCA_______ .......((.((((((((((.((((((((....))))))...)).))))......)))))).))...................................... (-21.88 = -21.99 + 0.11)


| Location | 1,941,789 – 1,941,884 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Shannon entropy | 0.40809 |
| G+C content | 0.51793 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.27 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1941789 95 - 27905053 -------UUGGAGCUGGGAGCUCGGAGUUCGAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU -------....(((((((...(((...(((....))).))))))((((....))))((..(((((((((....))))))(((....)))))).))...)))) ( -30.10, z-score = -0.34, R) >droSim1.chr3R 1969375 102 - 27517382 UUGGAGCUGGGAGCUGGGAGCUCGGAGUUCCAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU ((((((((..((((.....))))..))))))))((.......))((((....)))).((((..((((((....))))))(((....))).........)))) ( -38.60, z-score = -1.54, R) >droSec1.super_6 2043082 102 - 4358794 UUGGAGCUGGGAGCUGGGAGCUCGGAGUUCCAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU ((((((((..((((.....))))..))))))))((.......))((((....)))).((((..((((((....))))))(((....))).........)))) ( -38.60, z-score = -1.54, R) >droYak2.chr3R 17885077 102 - 28832112 UUGGAGCUGGGAGCUGGGAGCUCGGAGUUCCAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU ((((((((..((((.....))))..))))))))((.......))((((....)))).((((..((((((....))))))(((....))).........)))) ( -38.60, z-score = -1.54, R) >droAna3.scaffold_13340 5503461 86 + 23697760 ----------------UGAAGCAACGGCUCCAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU ----------------...(((..((.(((....))).)).((.((((....)))).)).(((((((((....))))))(((....)))))).......))) ( -26.70, z-score = -0.75, R) >dp4.chr2 14929321 92 + 30794189 ----------UGGGACUGGGACUGGAGCUGCAAGUAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU ----------........(..(((.((((((..........((.((((....)))).))....((((((....)))))))).))).).)))..)........ ( -28.50, z-score = -0.33, R) >droWil1.scaffold_181130 5301908 92 + 16660200 ----------CGUAGCCAGAGUUGGAGAUGCAAGUAGAUGACCCGCAUGAAAAUGCUGGCUCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU ----------...(((((...(((....((((.(((((.(.((.((((....)))).))))))((((((....))))))....)).))))...))).))))) ( -33.40, z-score = -2.48, R) >droMoj3.scaffold_6540 11216486 99 - 34148556 ---UGCUAGUUGUAGCUGGAACUGUAGCUGGGAGUAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUCCAGGACAAAUGGCU ---.((((.((((..(((((..(((.((.(((.........((.((((....)))).))))).((((((....))))))))..)))))))).)))).)))). ( -35.60, z-score = -1.90, R) >droGri2.scaffold_14624 1306050 96 + 4233967 ------GGAAACUGGGCAACUGUAUAGCUGGGAGUAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU ------(....)..(((..((((((.((((((.........((.((((....)))).))))).((((((....))))))...)))))))))..(....)))) ( -30.00, z-score = -0.66, R) >consensus _______UGGGAGCUGGGAGCUCGGAGCUCCAAGGAGAUGACCCGCAUGAAAAUGCUGGCCCUGGCAUAUUUUUAUGCCGCAAGCAUGCAGGACAAAUGGCU .........................((((............((.((((....)))).)).(((((((((....))))))(((....))))))......)))) (-21.44 = -21.27 + -0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:56 2011