| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,887,174 – 1,887,260 |
| Length | 86 |
| Max. P | 0.754520 |

| Location | 1,887,174 – 1,887,260 |
|---|---|
| Length | 86 |
| Sequences | 12 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 92.91 |
| Shannon entropy | 0.15611 |
| G+C content | 0.40317 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -18.06 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1887174 86 - 27905053 UGUGUGCGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCCCCAACUACGCUC .(((((.(((......)))........(((((((.((((((...(((..--..)))...)))))).)))))))........))))).. ( -17.30, z-score = -0.86, R) >droYak2.chr3R 17830936 86 - 28832112 UGUGUGCGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCCCCAACUACGCUA .(((((.(((......)))........(((((((.((((((...(((..--..)))...)))))).)))))))........))))).. ( -17.30, z-score = -0.71, R) >droEre2.scaffold_4770 2145341 85 - 17746568 UGUGUGCGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCCCCAACUACGCU- .(((((.(((......)))........(((((((.((((((...(((..--..)))...)))))).)))))))........))))).- ( -17.30, z-score = -0.82, R) >droSec1.super_6 1988604 86 - 4358794 UGUGUGCGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCCCCAACUACGCUC .(((((.(((......)))........(((((((.((((((...(((..--..)))...)))))).)))))))........))))).. ( -17.30, z-score = -0.86, R) >droSim1.chr3R 1914123 86 - 27517382 UGUGUGCGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCCCCAACUACGCUC .(((((.(((......)))........(((((((.((((((...(((..--..)))...)))))).)))))))........))))).. ( -17.30, z-score = -0.86, R) >droAna3.scaffold_13340 5452984 85 + 23697760 UGUGUGUGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCCCCAACCACACU- .(((((((((......)))).......(((((((.((((((...(((..--..)))...)))))).)))))))........))))).- ( -20.20, z-score = -2.27, R) >dp4.chr2 14881333 86 + 30794189 UGUGUGUGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCACCAACCACACAC ((((((((((......))))...((..(((((((.((((((...(((..--..)))...)))))).)))))))..))....)))))). ( -22.30, z-score = -1.99, R) >droPer1.super_0 4499479 79 - 11822988 UGUGUGUGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCACCAAC------- ..((.((((.......(((....))).(((((((.((((((...(((..--..)))...)))))).)))))))))))))..------- ( -18.10, z-score = -1.28, R) >droWil1.scaffold_181130 5241863 85 + 16660200 -GUAUGUGCACCUGUCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCACCAACCGCAUAC -(((((((((......)))....((..(((((((.((((((...(((..--..)))...)))))).)))))))..)).....)))))) ( -19.50, z-score = -1.68, R) >droVir3.scaffold_12822 2071063 86 + 4096053 UGUGUGCGCAGCUGCCUGCAAUCGUAAUUUCAAUAAAAGUAGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCACCAACCACACGC .(((((.((((....))))....((..(((((((.(((((....(((..--..)))....))))).)))))))..))....))))).. ( -21.20, z-score = -0.95, R) >droMoj3.scaffold_6540 11156390 86 - 34148556 UGUGUGUGCAGCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG--CACGUUGAAAUUUUAAUUGAAAGCACCAACCACACGC .((((((((((....)))))...((..(((((((.((((((...(((..--..)))...)))))).)))))))..))....))))).. ( -24.30, z-score = -1.98, R) >droGri2.scaffold_14624 1246419 88 + 4233967 UGUGUGUACAGCUGCUUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUGCACACGUUGAAAUUUUAAUUGAAAGCACCAACCACACGC .(((((....((.....))....((..(((((((.((((((...((((....))))...)))))).)))))))..))....))))).. ( -22.20, z-score = -1.23, R) >consensus UGUGUGCGCACCUGCCUGCAAUCGUAAUUUCAAUAAAAGUUGUUACGUG__CACGUUGAAAUUUUAAUUGAAAGCACCAACCACACUC .(((((.(((......)))........(((((((.((((((...((((....))))...)))))).)))))))........))))).. (-18.06 = -18.18 + 0.11)

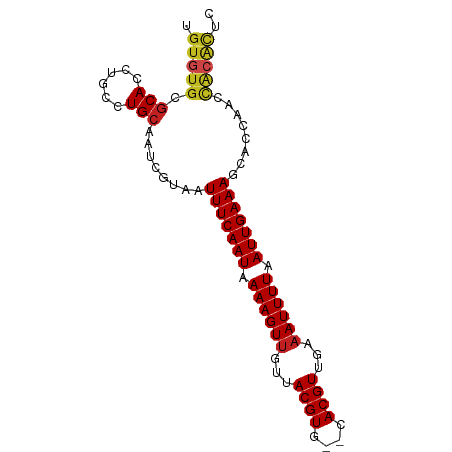

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:50 2011