| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,872,548 – 1,872,640 |
| Length | 92 |
| Max. P | 0.858626 |

| Location | 1,872,548 – 1,872,640 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.32394 |
| G+C content | 0.41123 |
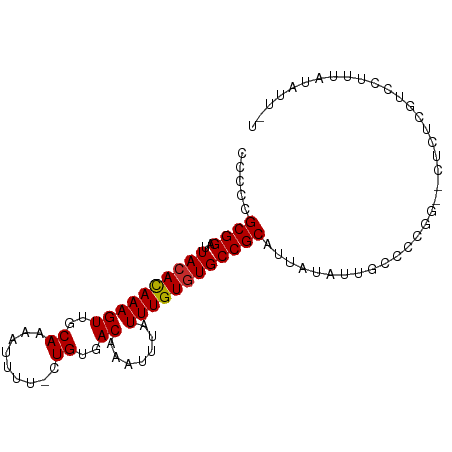
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
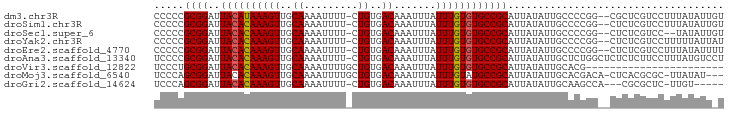
>dm3.chr3R 1872548 92 - 27905053 CCCCCGCGGAUUACAUAAAGUUGCAAAAUUUU-CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCCCCGG--CGCUCGUCCUUUAUAUUGU .......((((........((..((.......-.))..))..........(.((((((((......)))...))--))).))))).......... ( -19.50, z-score = -1.15, R) >droSim1.chr3R 1899185 92 - 27517382 CCCCCGCGGAUUACACAAAGUUGCAAAAUUUU-CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCCCCGG--CUCUCGUCCUUUAUAUUGU .....((((..((((((((((..((.......-.))..)).......))))))))))))........((....)--).................. ( -19.11, z-score = -1.54, R) >droSec1.super_6 1973676 90 - 4358794 CCCCCGCGGAUUACACAAAGUUGCAAAAUUUU-CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCCCCGG--CUCUCGUCC--UAUAUUGU .....((((..((((((((((..((.......-.))..)).......))))))))))))........((....)--)........--........ ( -19.11, z-score = -1.59, R) >droYak2.chr3R 17816351 92 - 28832112 CCCCCGCGGAUUACACAAAGUUGCAAAAUUUU-CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCCCCGG--CUCUCGUCCUUUUUAUUAU .....((((..((((((((((..((.......-.))..)).......))))))))))))........((....)--).................. ( -19.11, z-score = -1.90, R) >droEre2.scaffold_4770 2129466 92 - 17746568 CCCCCGCGGAUUACACAAAGUUGCAAAAUUUU-CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCCCCGG--CUCUCGUCCUUUAUAUUUU .....((((..((((((((((..((.......-.))..)).......))))))))))))........((....)--).................. ( -19.11, z-score = -1.90, R) >droAna3.scaffold_13340 5440310 94 + 23697760 UCCCCGCGGAUUACACAAAGUUGCAAAAUUUU-CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCUCUGGCUCUCUCUUCCUUUAUGUCCU .......((((.(((((((((..((.......-.))..)).......)))))))((((((......)))...)))...............)))). ( -18.51, z-score = -1.99, R) >droVir3.scaffold_12822 2056694 72 + 4096053 UCCCUGCGGAUUACACAAAGUUGCAAAAUUUUGCUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCACG----------------------- ....(((((..((((((((((..((.........))..)).......)))))))))))))............----------------------- ( -16.71, z-score = -1.21, R) >droMoj3.scaffold_6540 11144344 90 - 34148556 UCCCAGCGGAUUACACAAAGUUGCAAAAUUUUGCUGUGACAAAUUUAUUUGUAUGCCGCAUUAUAUUGCACGACA-CUCACGCGC-UUAUAU--- .....((((..(((((((((((....))))))).))))(((((....)))))...))))..((((..((.((...-....)).))-.)))).--- ( -19.30, z-score = -1.11, R) >droGri2.scaffold_14624 1233911 85 + 4233967 UCCCAGCGGAUUACACAAAGUUGCAAAAUUUU-CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCAAGCCA---CGCGCUC-UUGU----- .....((((..((((((((((..((.......-.))..)).......))))))))))))........(((((...---......)-))))----- ( -18.51, z-score = -0.45, R) >consensus CCCCCGCGGAUUACACAAAGUUGCAAAAUUUU_CUGUGACAAAUUUAUUUGUGUGCCGCAUUAUAUUGCCCCGG__CUCUCGUCCUUUAUAUU_U .....((((..((((((((((..((.........))..)).......)))))))))))).................................... (-15.65 = -15.67 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:47 2011