| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,864,705 – 1,864,814 |
| Length | 109 |
| Max. P | 0.774854 |

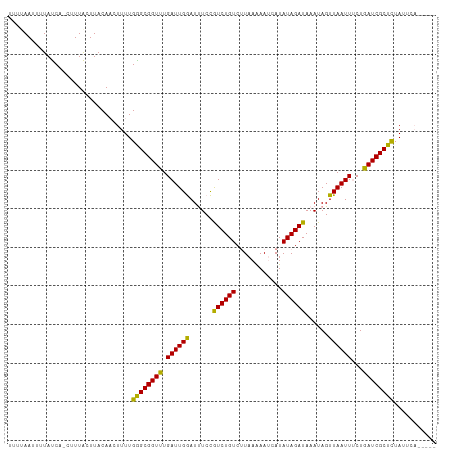
| Location | 1,864,705 – 1,864,812 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.45 |
| Shannon entropy | 0.21283 |
| G+C content | 0.28831 |
| Mean single sequence MFE | -15.67 |
| Consensus MFE | -15.90 |
| Energy contribution | -14.80 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.70 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1864705 107 + 27905053 UUUUAAUUUUACCAACUUUGCUUACAUCUUUUGGGCGGUUUGAUUGAAUCUCUAUCUGUCCUAAGAACUAUAUAGAUAAAUAGUUAAUUUCUAAUCGCUUUAUUCAC---- ...........((((...((....))....))))((((((.((((((.....(((((((............))))))).....))))))...)))))).........---- ( -15.80, z-score = -1.08, R) >droSim1.chr3R 1891469 110 + 27517382 UUUUAAUUUUAUCA-CUUUACUUACAACUUUUGGGCGGUUUGAUUGGAUUUCCGUCUGUCUUAAAAAUCAUAUAGAUAAAUAGUUAAUUUCUGAUCGCUCUAUUCAAUCGA ..............-.................((((((((.((((((......((((((............))))))......))))))...))))))))........... ( -15.50, z-score = -0.31, R) >droSec1.super_6 1966007 102 + 4358794 UUUAAAUUUUAUCA-CUUUACUUACAACUUUUGAGCGGUUUGAUUGGAUUUCCGUCUGUCUUAAAAAUCAUAUAGAUAAAUAGUUAAUUGCUGAUCGCUCUAU-------- ..............-.................((((((((.((((((......((((((............))))))......))))))...))))))))...-------- ( -15.70, z-score = -0.71, R) >consensus UUUUAAUUUUAUCA_CUUUACUUACAACUUUUGGGCGGUUUGAUUGGAUUUCCGUCUGUCUUAAAAAUCAUAUAGAUAAAUAGUUAAUUUCUGAUCGCUCUAUUCA_____ ................................((((((((.((((((......((((((............))))))......))))))...))))))))........... (-15.90 = -14.80 + -1.10)


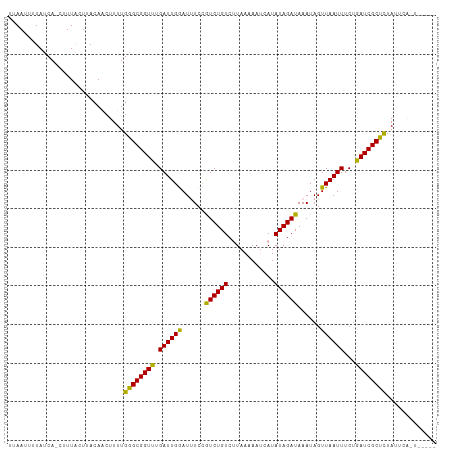
| Location | 1,864,707 – 1,864,814 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.93 |
| Shannon entropy | 0.23538 |
| G+C content | 0.29021 |
| Mean single sequence MFE | -15.67 |
| Consensus MFE | -15.90 |
| Energy contribution | -14.80 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.71 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1864707 107 + 27905053 UUAAUUUUACCAACUUUGCUUACAUCUUUUGGGCGGUUUGAUUGAAUCUCUAUCUGUCCUAAGAACUAUAUAGAUAAAUAGUUAAUUUCUAAUCGCUUUAUUCACUU---- .........((((...((....))....))))((((((.((((((.....(((((((............))))))).....))))))...))))))...........---- ( -15.80, z-score = -1.09, R) >droSim1.chr3R 1891471 110 + 27517382 UUAAUUUUAUCA-CUUUACUUACAACUUUUGGGCGGUUUGAUUGGAUUUCCGUCUGUCUUAAAAAUCAUAUAGAUAAAUAGUUAAUUUCUGAUCGCUCUAUUCAAUCGAUU ............-.................((((((((.((((((......((((((............))))))......))))))...))))))))............. ( -15.50, z-score = -0.24, R) >droSec1.super_6 1966009 100 + 4358794 UAAAUUUUAUCA-CUUUACUUACAACUUUUGAGCGGUUUGAUUGGAUUUCCGUCUGUCUUAAAAAUCAUAUAGAUAAAUAGUUAAUUGCUGAUCGCUCUAU---------- ............-.................((((((((.((((((......((((((............))))))......))))))...))))))))...---------- ( -15.70, z-score = -0.80, R) >consensus UUAAUUUUAUCA_CUUUACUUACAACUUUUGGGCGGUUUGAUUGGAUUUCCGUCUGUCUUAAAAAUCAUAUAGAUAAAUAGUUAAUUUCUGAUCGCUCUAUUCA_U_____ ..............................((((((((.((((((......((((((............))))))......))))))...))))))))............. (-15.90 = -14.80 + -1.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:46 2011