| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,863,151 – 1,863,258 |
| Length | 107 |
| Max. P | 0.599695 |

| Location | 1,863,151 – 1,863,258 |
|---|---|
| Length | 107 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Shannon entropy | 0.37938 |
| G+C content | 0.38771 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.79 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
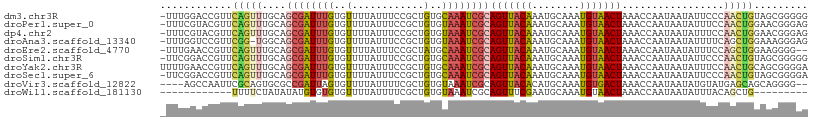
>dm3.chr3R 1863151 107 - 27905053 -UUUGGACCGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUGUGCAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUCCCAACUGUAGCGGGGG -......((((((((((....((((((((..(............)..))))))))(((((((........))))))).................))))).)))))... ( -30.50, z-score = -2.18, R) >droPer1.super_0 4477615 107 - 11822988 -UUUCGUACGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUGUGUAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUUCCAACUGGAACGGGAG -(..(((...(((((((....((((((((..(............)..))))))))(((((((........))))))).................))))))))))..). ( -25.80, z-score = -1.79, R) >dp4.chr2 14859737 107 + 30794189 -UUUCGUACGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUGUGUAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUUCCAACUGGAACGGGAG -(..(((...(((((((....((((((((..(............)..))))))))(((((((........))))))).................))))))))))..). ( -25.80, z-score = -1.79, R) >droAna3.scaffold_13340 5432519 106 + 23697760 -UUUGGUCCGUUCGG-UGGCAGCGAUUUGUGUUUUAUUUCCGCUGUGUAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUUUCAGCUGGAAAGGGAG -.....(((.(((((-(....((((((((..(............)..))))))))(((((((........)))))))..................))))))...))). ( -24.80, z-score = -0.67, R) >droEre2.scaffold_4770 2120144 105 - 17746568 -UUUGAACCGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUAUGCAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUUCCAGCUGGAAGGGG-- -......((.(((((((....(((((((((((............)))))))))))(((((((........))))))).................))))))).))..-- ( -28.20, z-score = -2.42, R) >droSim1.chr3R 1889836 107 - 27517382 -UUCGGACCGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUGUGCAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUCCCAACUGUAGCGGGGG -......((((((((((....((((((((..(............)..))))))))(((((((........))))))).................))))).)))))... ( -30.50, z-score = -2.29, R) >droYak2.chr3R 17801134 108 - 28832112 UUUUGAACCGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUGUGCAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUUCCAACUGCAGCGGGGA .......((((((((((....((((((((..(............)..))))))))(((((((........))))))).................))))).)))))... ( -30.50, z-score = -2.41, R) >droSec1.super_6 1964423 107 - 4358794 -UUCGGACCGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUGUGCAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUCCCAACUGUAGCGGGGA -......((((((((((....((((((((..(............)..))))))))(((((((........))))))).................))))).)))))... ( -30.50, z-score = -2.35, R) >droVir3.scaffold_12822 2045935 102 + 4096053 ----AGCCAAUUCGCAGUGCGCCGAUUAGUGUUUUAUUUUCGCUGUGUAAAUCGCAGUUACACAUGCAAAUGUGACUAAACCAAUAAUAUGUAUGAGCAGCAGGGG-- ----..((.....((.(((((...((((.((.((((.....((((((.....))))))..(((((....)))))..)))).)).)))).)))))..)).....)).-- ( -20.90, z-score = 1.19, R) >droWil1.scaffold_181130 5216129 87 + 16660200 ------------UUUUCUAUAUAUGUGUGUGUUUUAUUUUCGCUGUGUAAAUCGCAGUUUCGAAUGCAAAUGUAACUAAACCAAUAAUAUUUACAGCUG--------- ------------.......(((((...(((((((.......((((((.....))))))...))))))).))))).........................--------- ( -12.00, z-score = 0.28, R) >consensus _UUUGGACCGUUCAGUUUGCAGCGAUUUGUGUUUUAUUUCCGCUGUGCAAAUCGCAGUUACAAAUGCAAAUGUAACUAAACCAAUAAUAUUUCCAACUGGAACGGGGG ............(((((....((((((((..(............)..))))))))(((((((........))))))).................)))))......... (-16.58 = -16.79 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:44 2011