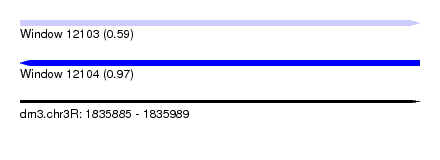
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,835,885 – 1,835,989 |
| Length | 104 |
| Max. P | 0.965919 |

| Location | 1,835,885 – 1,835,989 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.33 |
| Shannon entropy | 0.32302 |
| G+C content | 0.35340 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
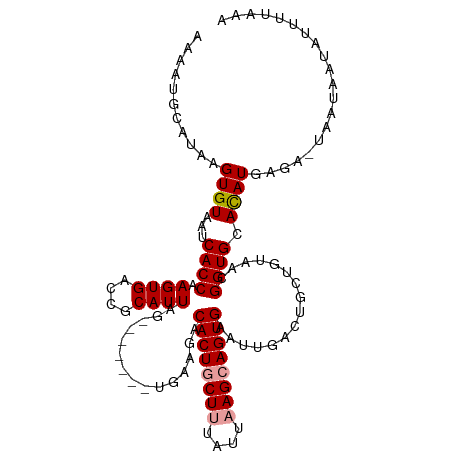
>dm3.chr3R 1835885 104 + 27905053 -AAAUAUAUAAGUGUGAUCACCAAGUGACCGCAUUAGCAAGCAGUAAAGGCACUGCUUUAUCUAGCAGUG---------UUAUACAGGUGCAUAUGAGA-UGGCAAUACUUUAAA -........((((((..((((((.((((((((....)).....(((..(((((((((......)))))))---------)).))).))).))).)).).-)))..)))))).... ( -27.90, z-score = -1.59, R) >droSec1.super_6 1945189 108 + 4358794 AAAAUGCAUAAGUGUAAUCACCAAGUGACCGCAUUAG-------AGAAGACACUGCUUUAUUAAGCAGUGAAUUGACUGCUGUAACGGUGGACAUGAGACUUAUAAUAUUUUAAA ((((((.((((((.(..(((((..((.((.(((...(-------(.....((((((((....))))))))..))...))).)).))))))).....).))))))..))))))... ( -25.10, z-score = -2.04, R) >droSim1.chr3R 1869993 108 + 27517382 AAGAUGCAUAAGUGUAAUCACCAAGUGACCGCAUUAG-------UGAAAACACUACUUUAUUAAGCAGUGAAUUGACUGCUGUAACGGUGCACAUGAGAUUAAUAAUAUUUUAAA ....(((((.(((((..((((..((((....)))).)-------)))..))))).........((((((......))))))......)))))..((((((.......)))))).. ( -23.70, z-score = -1.60, R) >consensus AAAAUGCAUAAGUGUAAUCACCAAGUGACCGCAUUAG_______UGAAGACACUGCUUUAUUAAGCAGUGAAUUGACUGCUGUAACGGUGCACAUGAGA_UAAUAAUAUUUUAAA ...........((((...((((.((((....))))...............((((((((....))))))))................)))).)))).................... (-16.19 = -16.63 + 0.45)

| Location | 1,835,885 – 1,835,989 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
| Shannon entropy | 0.32302 |
| G+C content | 0.35340 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -15.54 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
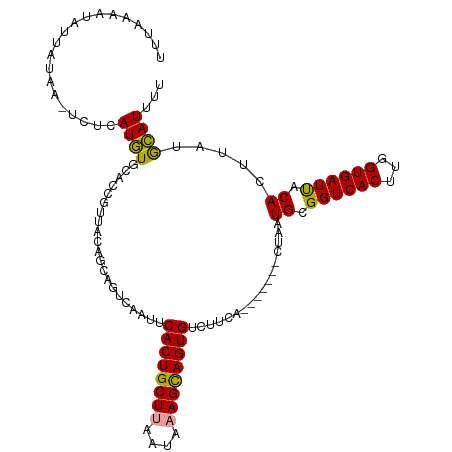
>dm3.chr3R 1835885 104 - 27905053 UUUAAAGUAUUGCCA-UCUCAUAUGCACCUGUAUAA---------CACUGCUAGAUAAAGCAGUGCCUUUACUGCUUGCUAAUGCGGUCACUUGGUGAUCACACUUAUAUAUUU- ......((((..(((-.....(((((....))))).---------.((((((((...((((((((....)))))))).)))..)))))....)))..)).))............- ( -26.70, z-score = -2.20, R) >droSec1.super_6 1945189 108 - 4358794 UUUAAAAUAUUAUAAGUCUCAUGUCCACCGUUACAGCAGUCAAUUCACUGCUUAAUAAAGCAGUGUCUUCU-------CUAAUGCGGUCACUUGGUGAUUACACUUAUGCAUUUU ..........(((((((.....(((.((((..((.(((.......((((((((....))))))))......-------....))).))....)))))))...)))))))...... ( -23.73, z-score = -2.60, R) >droSim1.chr3R 1869993 108 - 27517382 UUUAAAAUAUUAUUAAUCUCAUGUGCACCGUUACAGCAGUCAAUUCACUGCUUAAUAAAGUAGUGUUUUCA-------CUAAUGCGGUCACUUGGUGAUUACACUUAUGCAUCUU .............((((((((.(((.(((((......(((.((..((((((((....))))))))..)).)-------))...)))))))).))).))))).............. ( -26.70, z-score = -3.24, R) >consensus UUUAAAAUAUUAUAA_UCUCAUGUGCACCGUUACAGCAGUCAAUUCACUGCUUAAUAAAGCAGUGUCUUCA_______CUAAUGCGGUCACUUGGUGAUUACACUUAUGCAUUUU ....................((((.....................((((((((....)))))))).................((.((((((...)))))).)).....))))... (-15.54 = -14.77 + -0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:43 2011