| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,822,689 – 1,822,782 |
| Length | 93 |
| Max. P | 0.882109 |

| Location | 1,822,689 – 1,822,782 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.58723 |
| G+C content | 0.41353 |
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
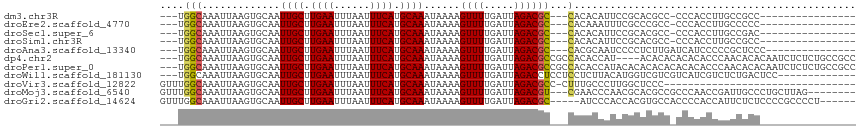
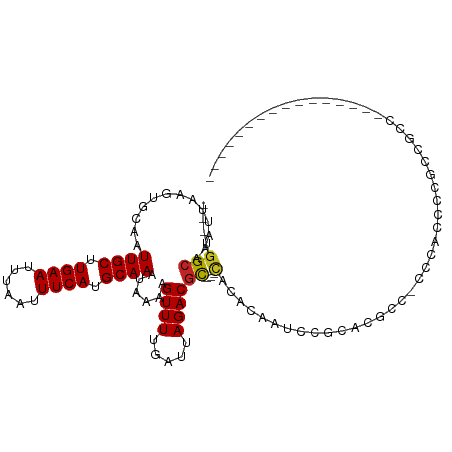
>dm3.chr3R 1822689 93 - 27905053 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGC---CACACAUUCCGCACGCC-CCCACCUUGCCGCC---------------- ---.(((((.....((((..((((.((((......)))).))))......((((.....))))..---..........))))...-......)))))...---------------- ( -20.24, z-score = -1.82, R) >droEre2.scaffold_4770 2087237 93 - 17746568 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGC---CACAAAUUUCGCCCGCC-CCCACCUUGCCCCC---------------- ---.(((((.......((..((((.((((......)))).))))......((((.....))))..---..............)).-......)))))...---------------- ( -15.94, z-score = -1.06, R) >droSec1.super_6 1931923 93 - 4358794 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGC---CACACAUUCCGCACGCC-CCCACCUUGCCGAC---------------- ---.(((((.....((((..((((.((((......)))).))))......((((.....))))..---..........))))...-......)))))...---------------- ( -20.24, z-score = -2.08, R) >droSim1.chr3R 1859233 93 - 27517382 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGC---CACACAUUCCGCACGCC-CCCACCUUGCCGCC---------------- ---.(((((.....((((..((((.((((......)))).))))......((((.....))))..---..........))))...-......)))))...---------------- ( -20.24, z-score = -1.82, R) >droAna3.scaffold_13340 5407638 95 + 23697760 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGC---CACGCAAUCCCCUCUUGAUCAUCCCCCGCUCCC--------------- ---((((.............((((.((((......)))).))))......((((.....))))))---))...............................--------------- ( -14.40, z-score = -0.45, R) >dp4.chr2 14831731 109 + 30794189 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGCCGCCACACCAU----ACACACACACACCCAACACACAAUCUCUCUGCCGCC ---.((((......(((...((((.((((......)))).))))......((((.....)))).............----..................)))........))))... ( -14.74, z-score = 0.05, R) >droPer1.super_0 4450969 113 - 11822988 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGCCGCCACACCAUACACACACACACACACCCAACACACAAUCUCUCUGCCGCC ---.((((......(((...((((.((((......)))).))))......((((.....))))...................................)))........))))... ( -14.74, z-score = 0.03, R) >droWil1.scaffold_181130 5190553 100 + 16660200 ---UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACCUCCUCCUCUUACAUGGUCGUCGUCAUCGUCUCUGACUCC------------- ---.(((.(((((((.....((((.((((......)))).))))........))))))).((((..............)))))))((((.......))))...------------- ( -18.36, z-score = -1.21, R) >droVir3.scaffold_12822 2012554 83 + 4096053 GUUUGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGCC-CUUUGCCCUUGGCUCCC-------------------------------- (((.(((((....((.((..((((.((((......)))).))))......((((.....)))))).-)))))))...)))....-------------------------------- ( -17.20, z-score = -0.98, R) >droMoj3.scaffold_6540 11102604 105 - 34148556 GUUUGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGU---CGAACCCAACGCACGCCGCCCAACCGAUUGCCCUGCUUAG-------- ((..(((((.....((((..((((.((((......)))).))))........((((((.....))---))))......))))..((......)).)))))..))....-------- ( -23.40, z-score = -1.58, R) >droGri2.scaffold_14624 1192524 105 + 4233967 GUUUGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGC-----AUCCCACCACGUGCCACCCCACCAUUCUCUCCCCGCCCCU------ ...(((((......((((..((((.((((......)))).))))......((((.....))))))-----)).........)))))........................------ ( -18.86, z-score = -2.03, R) >consensus ___UGGCAAAUUAAGUGCAAUUGCUUGAAUUUAAUUUCAUGCAAAUAAAAGUUUUGAUUAGACGC___CACACAAUCCGCACGCC_CCCACCCCGCCGCC________________ .....(((.......)))..((((.((((......)))).))))......((((.....))))..................................................... ( -9.72 = -9.72 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:41 2011