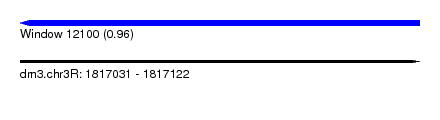
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,817,031 – 1,817,122 |
| Length | 91 |
| Max. P | 0.955506 |

| Location | 1,817,031 – 1,817,122 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Shannon entropy | 0.44721 |
| G+C content | 0.43881 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -20.94 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1817031 91 - 27905053 -UUUCCUCUUGCCAGCUGGCGGGGCUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAGGACAACUUGGUGGUCACGAUGGAAAAA------------------------- -((((((((((((....)))))))......................--.....((((((....(((((....)))))))))))....)))))..------------------------- ( -24.70, z-score = -1.56, R) >droSim1.chr3R 1853805 91 - 27517382 -UUUCCUCUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAGGACAACUUGGUGGUCACGAUGGAAAAA------------------------- -((((((((((((....)))))))......................--.....((((((....(((((....)))))))))))....)))))..------------------------- ( -24.80, z-score = -1.60, R) >droSec1.super_6 1926493 91 - 4358794 -UUUCCUCUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAGGACAACUGGGUGGUCACGAUGGAAAAA------------------------- -((((((((((((....)))))))......................--.....(((((((...((((.....)))))))))))....)))))..------------------------- ( -27.30, z-score = -2.05, R) >droYak2.chr3R 17763205 107 - 28832112 -UUUCCUCUUGCCAACUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUCUCCAAGACUACUUGGUGGUCACGAUGCUGUGCGCAGGAGGAAAAAAAA--------- -(((((((((((((...((((((((............)))).....--.....((((((....(((((....)))))))))))...)))).)).))))))))))).....--------- ( -35.40, z-score = -3.41, R) >droEre2.scaffold_4770 2082113 107 - 17746568 -UUUCCUCUUGCCAGCUGGCGGGGCUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGCUGCUGUGCGCAGGAGGAGAUGCUG--------- -(((((((((((((((.(((((((..............))).....--.....((((((....(((((....)))))))))))))))))))...))))))))))).....--------- ( -39.34, z-score = -3.27, R) >dp4.chr2 14827169 109 + 30794189 UGCUCCUUCUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGUUGCCGCCCAUUGAUGA--UGCGAGGGA------ ...((((((.(((((..(((((((((.......)))).........--.....((((((....(((((....))))))))))).....)))))..)))....--.))))))))------ ( -30.40, z-score = -0.48, R) >droPer1.super_0 4446696 109 - 11822988 UGCUCCUUCUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGUUGCCGCCCAUUGAUGA--UGCGAGGGA------ ...((((((.(((((..(((((((((.......)))).........--.....((((((....(((((....))))))))))).....)))))..)))....--.))))))))------ ( -30.40, z-score = -0.48, R) >droAna3.scaffold_13340 5402748 106 + 23697760 UUUUCCUCUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUUUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGAUUUAGCUCAUUGAGGAAAAG------------- ((((((((.....(((((.((((((............))))............((((((....(((((....)))))))))))))...)))))....)))))))).------------- ( -29.60, z-score = -2.84, R) >droWil1.scaffold_181130 5185178 113 + 16660200 -CUUGCUUUUGCCAGCUGGCGGAGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGUU---GCCCAUUGAUGAUGUGCCAGUGAGAAGAG -....(((((.(..(((((((((((....)))).((((((......--.....((((((....(((((....)))))))))))....---.......)))))).)))))))..)))))) ( -31.55, z-score = -1.72, R) >droVir3.scaffold_12822 2006251 108 + 4096053 -CAUGCCUUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGUUG--GUCCAUUGAUGA--UGCCAGAAGAA---- -.....(((((((....)))))))(((((.....((((((.((..(--((...((((((....(((((....)))))))))))....)--))..)).)))))--)....))))).---- ( -28.50, z-score = -0.58, R) >droMoj3.scaffold_6540 11097068 108 - 34148556 -UUUGCCUUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCAUGUUG--GCCCAUUGAUGA--UGCCAGAAAAA---- -.....(((((((....)))))))(((((.....((((((......--.....((((((....(((((....))))))))))).....--.......)))))--)....))))).---- ( -28.50, z-score = -0.54, R) >droGri2.scaffold_14624 1187428 107 + 4233967 -CUCGCCUUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA--UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCAUGUU---GCCCAUUGAUGA--UGCCAGAAGAA---- -((((((..........)))))).(((((.....((((((......--.....((((((....(((((....)))))))))))....---.......)))))--)....))))).---- ( -30.35, z-score = -1.37, R) >consensus _UUUCCUCUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUA__UUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGUUG__GCCCAUUGAUGA__UGCCAG_________ ......(((((((....))))))).............................((((((....(((((....))))))))))).................................... (-20.94 = -20.18 + -0.76)

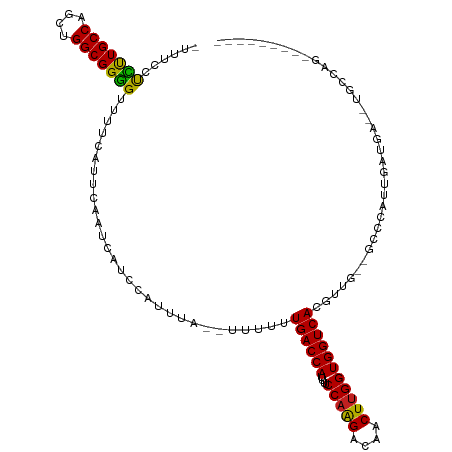
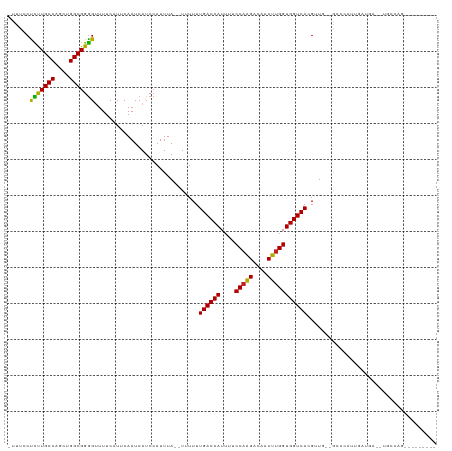
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:40 2011