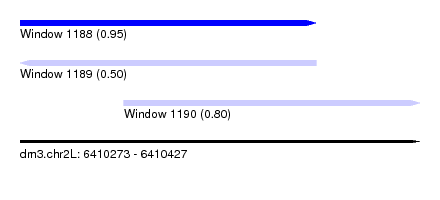
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,410,273 – 6,410,427 |
| Length | 154 |
| Max. P | 0.948001 |

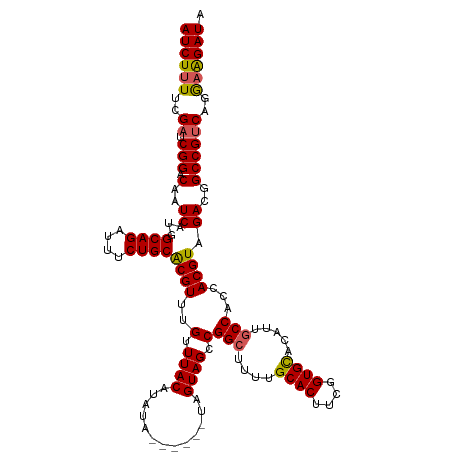
| Location | 6,410,273 – 6,410,387 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Shannon entropy | 0.10642 |
| G+C content | 0.47116 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -29.44 |
| Energy contribution | -32.56 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
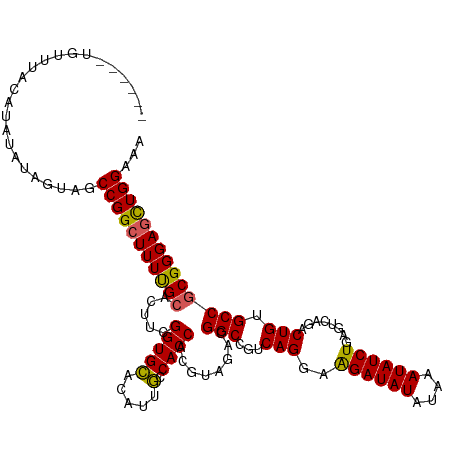
>dm3.chr2L 6410273 114 + 23011544 AUCUUUUCGAUCGGACAAUCAUUGCAGAUUUCUGCACGUUUGUUUACAUAUA------UAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUA ((((((..((.(((.(......(((((....)))))((((((.((((.....------..))))..(((...(((((....)))))....)))......))))))))))))..)))))). ( -37.90, z-score = -3.85, R) >droSim1.chr2L 6214696 114 + 22036055 AUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCGCGUUUGUUUACAUAUA------UAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUA ((((((..((.(((.(..((...((((....))))((((..(.((((.....------..)))).)(((...(((((....)))))....)))...)))).))..))))))..)))))). ( -37.40, z-score = -2.91, R) >droSec1.super_5 4477971 114 + 5866729 AUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCGCGUUUGUUUACAUAUA------UAGUAGCCGGCUUUUGCACUUCGGUGUUCAUUACCACCACGUAGACGGCCGUCAGGAAGAUA ((((((..((.(((......(((((((....)))).)))((((((((.....------.........((....)).....((((........))))..)))))))))))))..)))))). ( -31.50, z-score = -1.60, R) >droYak2.chr2L 15830603 114 - 22324452 AUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCACGUUUGUUUACAUAUA------UAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAGGAUA ((((((..((.(((.(.......((((....)))).((((((.((((.....------..))))..(((....((((....)))).....)))......))))))))))))..)))))). ( -35.40, z-score = -2.49, R) >droEre2.scaffold_4929 15336525 119 + 26641161 AUCUCUUCGAUCGGACAAUCAUGGCAGAUU-CUGCACGUUUGUUUACAUAUACGUGUAUAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGGCAGAAAGAUA ((((.(((((((...((....))...))))-.(((((((.((....))...))))))).....(((((((.((((((....))))....(((......))))).))))))).))))))). ( -39.30, z-score = -2.67, R) >consensus AUCUUUUCGAUCGGACAAUCAUGGCAGAUUUCUGCACGUUUGUUUACAUAUA______UAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUA ((((((..((.(((.........((((....))))....((((((((.(((.........)))...(((....((((....)))).....))).....)))))))))))))..)))))). (-29.44 = -32.56 + 3.12)

| Location | 6,410,273 – 6,410,387 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Shannon entropy | 0.10642 |
| G+C content | 0.47116 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.72 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
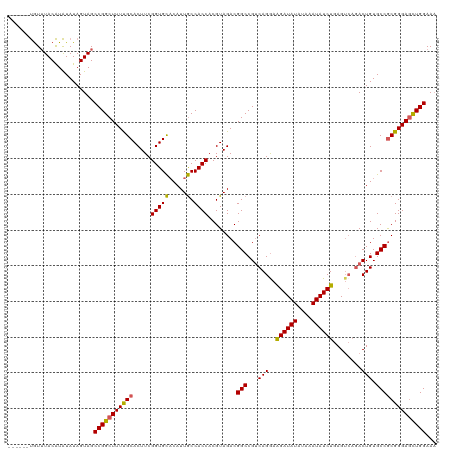
>dm3.chr2L 6410273 114 - 23011544 UAUCUUCCUGACGGCCGUCUACGUGGUGGCAAUGUGCACCGAAGUGCAAAAGCCGGCUACUA------UAUAUGUAAACAAACGUGCAGAAAUCUGCAAUGAUUGUCCGAUCGAAAAGAU .(((((..(((((((.(((...((((((((....(((((....)))))...))).)))))..------................(((((....)))))..))).).))).)))..))))) ( -34.00, z-score = -2.41, R) >droSim1.chr2L 6214696 114 - 22036055 UAUCUUCCUGACGGCCGUCUACGUGGUGGCAAUGUGCACCGAAGUGCAAAAGCCGGCUACUA------UAUAUGUAAACAAACGCGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAU .(((((..(((((((.(((...((((((((....(((((....)))))...))).)))))..------...............(.((((....)))))..))).).))).)))..))))) ( -32.90, z-score = -1.79, R) >droSec1.super_5 4477971 114 - 5866729 UAUCUUCCUGACGGCCGUCUACGUGGUGGUAAUGAACACCGAAGUGCAAAAGCCGGCUACUA------UAUAUGUAAACAAACGCGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAU .(((((..(((((((.((((((((((((........))))..(((((........)).))).------...))))).......(.((((....)))))..))).).))).)))..))))) ( -27.30, z-score = -0.78, R) >droYak2.chr2L 15830603 114 + 22324452 UAUCCUCCUGACGGCCGUCUACGUGGUGGCAAUGUGCACCGAAGUGCGAAAGCCGGCUACUA------UAUAUGUAAACAAACGUGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAU ...........((..((.....((((((((....(((((....)))))...))).)))))..------.........((((.(((((((....))).)))).)))).))..))....... ( -29.30, z-score = -0.71, R) >droEre2.scaffold_4929 15336525 119 - 26641161 UAUCUUUCUGCCGGCCGUCUACGUGGUGGCAAUGUGCACCGAAGUGCGAAAGCCGGCUACUAUACACGUAUAUGUAAACAAACGUGCAG-AAUCUGCCAUGAUUGUCCGAUCGAAGAGAU .(((((((.((((((..(((((.(((((........)))))..))).))..))))))(((((((....)))).))).((((.(((((((-...))).)))).))))......).)))))) ( -33.60, z-score = -0.72, R) >consensus UAUCUUCCUGACGGCCGUCUACGUGGUGGCAAUGUGCACCGAAGUGCAAAAGCCGGCUACUA______UAUAUGUAAACAAACGUGCAGAAAUCUGCCAUGAUUGUCCGAUCGAAAAGAU .(((((..(((((((.(((...((((((((....(((((....)))))...))).))))).........................((((....))))...))).).))).)))..))))) (-28.40 = -28.72 + 0.32)

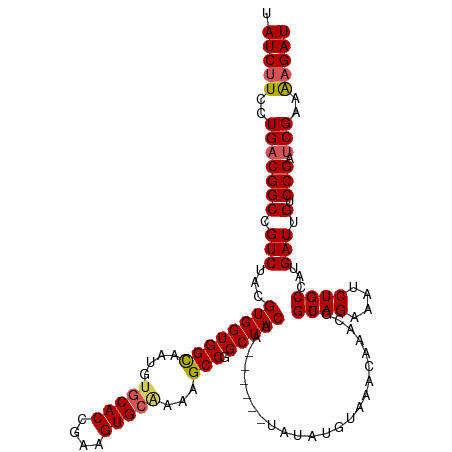
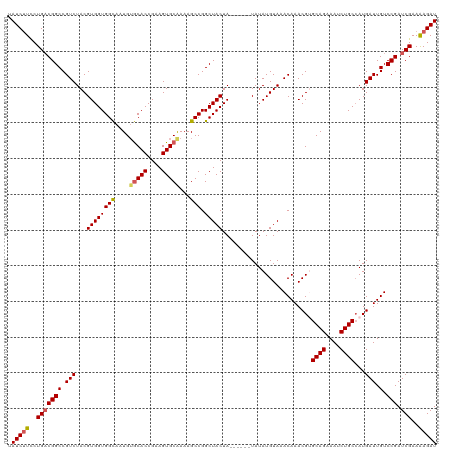
| Location | 6,410,313 – 6,410,427 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Shannon entropy | 0.16057 |
| G+C content | 0.47797 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -31.36 |
| Energy contribution | -31.48 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6410313 114 + 23011544 ------UGUUUACAUAUAUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAACUGGAAA ------............((((..(((((...(((((....)))))....)))........(.((((((((.((.((((((....))))))...)).))).).)))))))..)))).... ( -34.50, z-score = -1.38, R) >droSim1.chr2L 6214736 113 + 22036055 ------UGUUUACAUAUAUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAGCUGGAA- ------..................(((((((((((.....(((((.....).))))........(((((((.((.((((((....))))))...)).))).).))))))))))))))..- ( -36.30, z-score = -1.52, R) >droSec1.super_5 4478011 110 + 5866729 ------UGUUUACAUAUAUAGUAGCCGGCUUUUGCACUUCGGUGUUCAUUACCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAG----ACUGUGCCGCGGGAGCUGGAAA ------..................(((((((((((.....((((........))))........(((...(((..((((((....))))))...----.))).))))))))))))))... ( -35.40, z-score = -2.22, R) >droYak2.chr2L 15830643 114 - 22324452 ------UGUUUACAUAUAUAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAGGAUAUAUAAAUAUCUGAGUCAGACUGUGCCACGGGAGUUGGAAA ------..................((((((..(((((....))))...................(((((((.((.((((((....))))))...)).))).).)))..)..))))))... ( -33.70, z-score = -0.96, R) >droEre2.scaffold_4929 15336564 120 + 26641161 UGUUUACAUAUACGUGUAUAGUAGCCGGCUUUCGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGGCAGAAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAGUUGGAAA ......((....((((((((((.(((((((.((((((....))))....(((......))))).))))))).((.((((((....))))))...))..)))))).)))).....)).... ( -42.30, z-score = -2.63, R) >consensus ______UGUUUACAUAUAUAGUAGCCGGCUUUUGCACUUCGGUGCACAUUGCCACCACGUAGACGGCCGUCAGGAAGAUAUAUAAAUAUCUGAGUCAGACUGUGCCGCGGGAGCUGGAAA ........................(((((((((((.....(((((.....).))))........(((((((.((.((((((....))))))...)).))).).))))))))))))))... (-31.36 = -31.48 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:15 2011