
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,791,834 – 1,791,931 |
| Length | 97 |
| Max. P | 0.987294 |

| Location | 1,791,834 – 1,791,931 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.37 |
| Shannon entropy | 0.22304 |
| G+C content | 0.35290 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
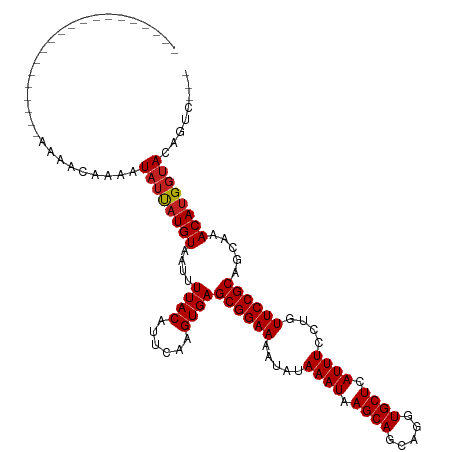
>dm3.chr3R 1791834 97 + 27905053 --------------------AAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUC--- --------------------.........((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))))).....--- ( -21.40, z-score = -2.36, R) >droSim1.chr3R 1827103 97 + 27517382 --------------------AAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUC--- --------------------.........((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))))).....--- ( -21.40, z-score = -2.36, R) >droSec1.super_6 1901568 97 + 4358794 --------------------AAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUC--- --------------------.........((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))))).....--- ( -21.40, z-score = -2.36, R) >droYak2.chr3R 17737497 104 + 28832112 -------------AAAAAGAAAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUC--- -------------................((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))))).....--- ( -21.40, z-score = -2.28, R) >droEre2.scaffold_4770 2057478 97 + 17746568 --------------------AAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUC--- --------------------.........((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))))).....--- ( -21.40, z-score = -2.36, R) >droAna3.scaffold_13340 5379443 97 - 23697760 --------------------AAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACACGGUACAGUC--- --------------------.........(((..(((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))..))).....--- ( -19.10, z-score = -1.68, R) >dp4.chr2 14802335 100 - 30794189 --------------------ACGAAAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGCCAGC --------------------.........((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))))))........ ( -21.40, z-score = -1.28, R) >droPer1.super_0 4420471 100 + 11822988 --------------------ACGAAAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGCCAGC --------------------.........((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))))))........ ( -21.40, z-score = -1.28, R) >droWil1.scaffold_181130 5157655 98 - 16660200 ---------------------AAAAAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUCAG- ---------------------........((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))))).......- ( -21.40, z-score = -2.36, R) >droVir3.scaffold_12822 1977011 115 - 4096053 -ACAAGAAUAAAUAAAAAUAAAGAGAAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACACU---- -............................((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))))))....---- ( -23.50, z-score = -2.54, R) >droMoj3.scaffold_6540 11068404 115 + 34148556 UACAAGAA-AUAUAUAUAUAAAGAGAAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGCACAGU---- ........-......................((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))))......---- ( -20.80, z-score = -1.21, R) >droGri2.scaffold_14624 1162417 101 - 4233967 ---------------AAGAACCGAGAAUAUAACAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGCUACAGU---- ---------------.................(((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))).......---- ( -19.90, z-score = -0.94, R) >consensus ____________________AAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUC___ .............................((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))))))........ (-21.22 = -21.28 + 0.06)
| Location | 1,791,834 – 1,791,931 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.37 |
| Shannon entropy | 0.22304 |
| G+C content | 0.35290 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.48 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
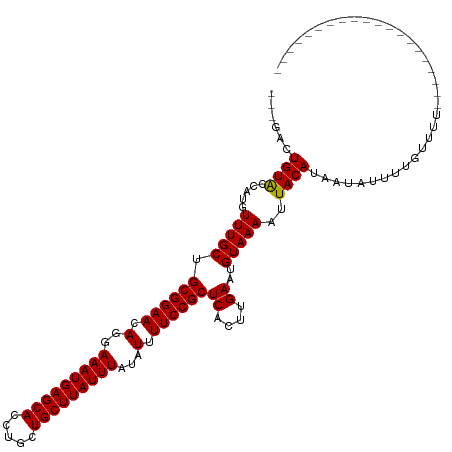
>dm3.chr3R 1791834 97 - 27905053 ---GACUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUU-------------------- ---...((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..))))..............-------------------- ( -20.50, z-score = -1.81, R) >droSim1.chr3R 1827103 97 - 27517382 ---GACUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUU-------------------- ---...((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..))))..............-------------------- ( -20.50, z-score = -1.81, R) >droSec1.super_6 1901568 97 - 4358794 ---GACUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUU-------------------- ---...((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..))))..............-------------------- ( -20.50, z-score = -1.81, R) >droYak2.chr3R 17737497 104 - 28832112 ---GACUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUUUCUUUUU------------- ---...((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..)))).....................------------- ( -20.50, z-score = -1.77, R) >droEre2.scaffold_4770 2057478 97 - 17746568 ---GACUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUU-------------------- ---...((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..))))..............-------------------- ( -20.50, z-score = -1.81, R) >droAna3.scaffold_13340 5379443 97 + 23697760 ---GACUGUACCGUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUU-------------------- ---.........(((......((((((.(..(((((((((.....)))))))))...).)))))).)))....(((((....))))).............-------------------- ( -21.00, z-score = -1.77, R) >dp4.chr2 14802335 100 + 30794189 GCUGGCUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUUUCGU-------------------- ((..((........))..)).((((((.(..(((((((((.....)))))))))...).))))))........(((((....))))).............-------------------- ( -23.00, z-score = -1.53, R) >droPer1.super_0 4420471 100 - 11822988 GCUGGCUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUUUCGU-------------------- ((..((........))..)).((((((.(..(((((((((.....)))))))))...).))))))........(((((....))))).............-------------------- ( -23.00, z-score = -1.53, R) >droWil1.scaffold_181130 5157655 98 + 16660200 -CUGACUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUUUUU--------------------- -.....((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..)))).............--------------------- ( -20.50, z-score = -2.11, R) >droVir3.scaffold_12822 1977011 115 + 4096053 ----AGUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUGAUAUUUUCUCUUUAUUUUUAUUUAUUCUUGU- ----.(((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..)))))................................- ( -22.20, z-score = -1.51, R) >droMoj3.scaffold_6540 11068404 115 - 34148556 ----ACUGUGCCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUGAUAUUUUCUCUUUAUAUAUAUAU-UUCUUGUA ----.......(((((((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..))))....))))).......................-........ ( -22.10, z-score = -1.37, R) >droGri2.scaffold_14624 1162417 101 + 4233967 ----ACUGUAGCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUGUUAUAUUCUCGGUUCUU--------------- ----..((((((((((((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..))))....))))))))))............--------------- ( -27.70, z-score = -3.00, R) >consensus ___GACUGUACCAUGUUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUU____________________ ......((((.....(((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..)))).................................. (-20.56 = -20.48 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:37 2011