
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,790,373 – 1,790,466 |
| Length | 93 |
| Max. P | 0.656691 |

| Location | 1,790,373 – 1,790,466 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.89 |
| Shannon entropy | 0.48314 |
| G+C content | 0.43068 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.12 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1790373 93 - 27905053 --------------------UCUCGCCGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACGAGGGAGGGGAG --------------------.(((.((.......((((((((.....))))))))....((..(((((......)))))((((........))))........))..)).))) ( -20.60, z-score = -1.19, R) >droSim1.chr3R 1825642 85 - 27517382 --------------------UCUCGCCGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACGGAG-------- --------------------.....(((.((...((((((((.....))))))))...))...(((((......)))))((((........)))).....)))..-------- ( -17.00, z-score = -1.37, R) >droSec1.super_6 1900103 85 - 4358794 --------------------UCUCGCCGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACGGAG-------- --------------------.....(((.((...((((((((.....))))))))...))...(((((......)))))((((........)))).....)))..-------- ( -17.00, z-score = -1.37, R) >droYak2.chr3R 17736038 85 - 28832112 --------------------UCUGGCCAAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAGCGGAG-------- --------------------((((((((......((((((((.....))))))))...((.......)).....)))).((((........)))).....)))).-------- ( -16.30, z-score = -0.48, R) >droEre2.scaffold_4770 2056068 85 - 17746568 --------------------UCUCGCCGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACGGCG-------- --------------------...(((((.((...((((((((.....))))))))...))...(((((......)))))((((........)))).....)))))-------- ( -21.60, z-score = -2.68, R) >droAna3.scaffold_13340 5377722 102 + 23697760 UCUCGCCUUCGUCGU---UCCGUUGUCGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGAACAUAAUUUACGAUACGAACAGCAGAG-------- .(((((.(((((.((---((((((((......)).))))))))....((((((((...((.......))..)))))))).............)))))..)).)))-------- ( -23.20, z-score = -1.40, R) >dp4.chr2 14800340 105 + 30794189 UCUCGGUUCUGUUGCUCGUCGUCGGCAGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACGGAG-------- .......(((((((.((((.(((((((..((...((((((((.....))))))))...))...))))((.......))...........))))))).))))))).-------- ( -30.30, z-score = -2.56, R) >droPer1.super_0 4418496 105 - 11822988 UCUCGGUUCUGUUGCUCGUCGUCGUCAGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACGGAG-------- .......(((((((.((((.(((((..(.((...((((((((.....))))))))...)))..(((((......)))))........))))))))).))))))).-------- ( -29.50, z-score = -2.59, R) >droWil1.scaffold_181130 5155735 92 + 16660200 UCUCGCCUGUGUCGC----------UUGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCCAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACG----------- ....((((((((.((----------.((.((...((((((((.....))))))))...)).)).)).)))))...))).((((........)))).......----------- ( -22.20, z-score = -1.87, R) >droGri2.scaffold_14624 1159725 89 + 4233967 ----------------------UCUCGACGCAACAAAUGGGACAAAUUUCCAUUUCUAUCCAAUGCCGCAUAAAUGGCAAAUAAUUUACGAUA-CGAUAUGAGGCAGCAGCA- ----------------------.((((.......((((((((.....))))))))........(((((......)))))..............-.....))))((....)).- ( -15.40, z-score = -0.14, R) >droMoj3.scaffold_6540 11065368 90 - 34148556 ----------------------UCUCGACGCAACAAAUGGGACAAAUUUCCAUUUCUAUCCAAUGCUGCAUAAAUGGCACAUAAUUUACGAUACCGUUAUGUGGCAGCAGAG- ----------------------.(((...((...((((((((.....))))))))........(((..(((((.(((................))))))))..))))).)))- ( -21.59, z-score = -2.06, R) >droVir3.scaffold_12822 1974273 90 + 4096053 ----------------------UCUCGACGCAACAAAUGGGACAAAUUUCCAUUUCUAUCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUACCGUUAUGAGGCAGCGCAG- ----------------------....(.(((...((((((((.....))))))))........((((.(((((.(((................)))))))).))))))))..- ( -21.59, z-score = -2.10, R) >consensus ____________________UCUCGCCGAGCAACAAAUGGGACAAAUUUCCAUUUCAAGCCAAUGCCGCAUAAAUGGCACAUAAUUUACGAUAUGAACAACGGAG________ ..................................((((((((.....))))))))........(((((......))))).................................. (-10.12 = -10.12 + 0.01)

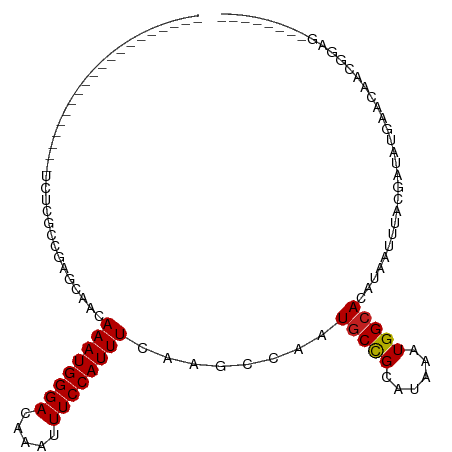
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:35 2011