| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,760,118 – 1,760,239 |
| Length | 121 |
| Max. P | 0.912512 |

| Location | 1,760,118 – 1,760,239 |
|---|---|
| Length | 121 |
| Sequences | 8 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 65.30 |
| Shannon entropy | 0.62708 |
| G+C content | 0.48201 |
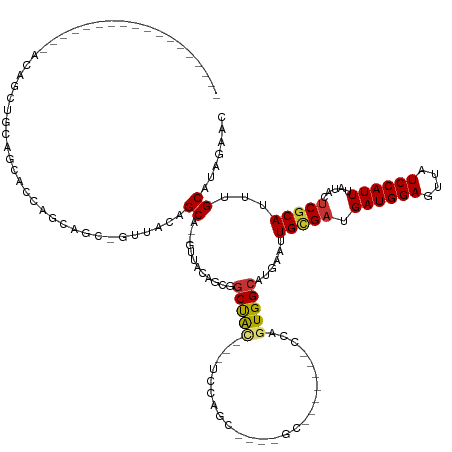
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -14.91 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
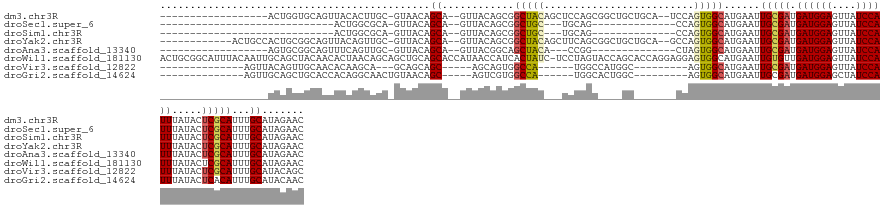
>dm3.chr3R 1760118 121 - 27905053 ------------------ACUGGUGCAGUUACACUUGC-GUAACAGCA--GUUACAGCGGCUACAGCUCCAGCGGCUGCUGCA--UCCAGUGGCAUGAAUUGCGAUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUAGAAC ------------------((((((((.(((((......-)))))((((--((..(.(.(((....))).).)..)))))))))--.))))).(((.....(((((.((((((....)))))).....)))))..)))....... ( -43.30, z-score = -2.17, R) >droSec1.super_6 1868422 95 - 4358794 -----------------------------ACUGGCGCA-GUUACAGCA--GUUACAGCGGCUGC---UGCAG--------------CCAGUGGCAUGAAUUGCGAUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUAGAAC -----------------------------(((((((((-(...((((.--((....)).)))))---))).)--------------))))).(((.....(((((.((((((....)))))).....)))))..)))....... ( -35.80, z-score = -2.80, R) >droSim1.chr3R 1794974 95 - 27517382 -----------------------------ACUGGCGCA-GUUACAGCA--GUUACAGCGGCUGC---UGCAG--------------CCAGUGGCAUGAAUUGCGAUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUAGAAC -----------------------------(((((((((-(...((((.--((....)).)))))---))).)--------------))))).(((.....(((((.((((((....)))))).....)))))..)))....... ( -35.80, z-score = -2.80, R) >droYak2.chr3R 17704396 127 - 28832112 ------------ACUGCCACUGCGGCAGUUACAGUUGC-GUUACAGCA--GUUACAGCGGCUACAGCUUCAGCGGCUGCUGCA--GCCAGUGGCAUGAAUUGCGAUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUAGAAC ------------...(((((((..((((......))))-(((.(((((--((..(.(.(((....))).).)..))))))).)--)))))))))(((...(((((.((((((....)))))).....)))))....)))..... ( -46.90, z-score = -1.84, R) >droAna3.scaffold_13340 5350582 106 + 23697760 ------------------AGUGCGGCAGUUUCAGUUGC-GUUACAGCA--GUUACGGCAGCUACA---CCGG--------------CUAGUGGCAUGAAUUGCGAUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUAGAAC ------------------((((((((((((.((..(((-......)))--(((((...((((...---..))--------------)).))))).))))))))...((((((....))))))......)))))).......... ( -29.50, z-score = -0.36, R) >droWil1.scaffold_181130 5120044 143 + 16660200 ACUGCGGCAUUUACAAUUGCAGCUACAACACUAACAGCAGCUGCAGCACCAUAACCAUCACUAUC-UCCUAGUACCAGCACCAGGAGGAGUGGCAUGAAUUGUGUUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUAGAAC ..((((((........((((((((.(..........).))))))))............((((.((-((((.((......)).)))))))))))).......((((.((((((....)))))).)))).))))............ ( -39.60, z-score = -1.66, R) >droVir3.scaffold_12822 1938999 107 + 4096053 --------------AGUUACAGUUGCAACACAAGCA---GCAGCAGC-----AGCAGUGGCCA------UGGCCAUGGC---------AGUGGCAUGAAUUGCGAUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUACAGC --------------.(((((.(((((.......)))---))......-----.((.(((((..------..))))).))---------.)))))(((...(((((.((((((....)))))).....)))))....)))..... ( -36.60, z-score = -1.71, R) >droGri2.scaffold_14624 1127512 110 + 4233967 --------------AGUUGCAGCUGCACCACAGGCAACUGUAACAGC-----AGUCGUGGCCA------UGGCACUGGC---------AGUGGCAUGAAUUGCGAUGAUGGAGCUAUCCAUUUAUACUCACAUUUGCAUACAAC --------------.(((((((.(((.......))).))))))).((-----(((((..(.((------((.(((....---------.))).))))..)..))))((((((....))))))............)))....... ( -36.70, z-score = -1.13, R) >consensus __________________ACAGCUGCAGCACCAGCAGC_GUUACAGCA__GUUACAGCGGCUAC___UCCAGC____GC_______CCAGUGGCAUGAAUUGCGAUGAUGGAGUUAUCCAUUUAUACUCGCAUUUGCAUAGAAC .......................................((.((.((.........)).((((......))))................)).))(((...(((((.((((((....)))))).....)))))....)))..... (-14.91 = -14.60 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:32 2011