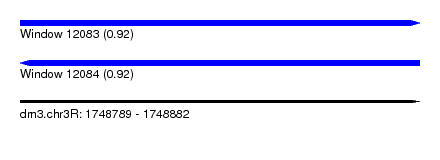
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,748,789 – 1,748,882 |
| Length | 93 |
| Max. P | 0.923040 |

| Location | 1,748,789 – 1,748,882 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Shannon entropy | 0.20568 |
| G+C content | 0.31811 |
| Mean single sequence MFE | -19.49 |
| Consensus MFE | -14.15 |
| Energy contribution | -15.26 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1748789 93 + 27905053 GUAUUCAAAAUACAUGCUCUCCUUUUAAUCAUCAUAAACCGUCAGCAUUCAGUAGUUGGUCUGAAUUGGUUUUUUUUCU----AAAGCAUGUGUUUC .......(((((((((((.................((((((.....((((((........)))))))))))).......----..))))))))))). ( -18.07, z-score = -2.82, R) >droSim1.chr3R 1782998 96 + 27517382 GUUUUCAAAAUACAUGCUGUUUUUUUAAUCAUCAUAGACCGUCAGC-GGAAUUAGUUGGUCUGAAUUUGUGUUUUUUUUUUCAAAAGCAUGUGUUUC .......(((((((((((..............(((((((((....)-))..((((.....)))).))))))..............))))))))))). ( -21.49, z-score = -2.62, R) >droSec1.super_6 1856829 94 + 4358794 GUUUUCAAAAGACAUGCUGUUCUUUUAAUUAUCAUAGACCGUCAGC-UGAAUUAGUUGGUCUAAGUUUGUAUUUUUUUU--CAAAAGCAUGUGUUUC .......(((.(((((((................(((((((...((-((...)))))))))))..((((..........--))))))))))).))). ( -18.90, z-score = -2.02, R) >consensus GUUUUCAAAAUACAUGCUGUUCUUUUAAUCAUCAUAGACCGUCAGC_UGAAUUAGUUGGUCUGAAUUUGUAUUUUUUUU__CAAAAGCAUGUGUUUC .......(((((((((((................(((((((...............)))))))..((((............))))))))))))))). (-14.15 = -15.26 + 1.11)

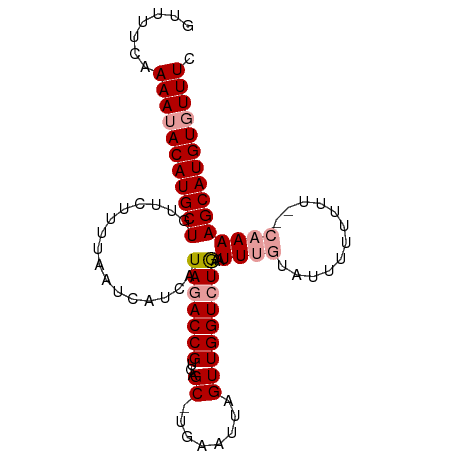
| Location | 1,748,789 – 1,748,882 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.07 |
| Shannon entropy | 0.20568 |
| G+C content | 0.31811 |
| Mean single sequence MFE | -16.17 |
| Consensus MFE | -12.22 |
| Energy contribution | -12.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1748789 93 - 27905053 GAAACACAUGCUUU----AGAAAAAAAACCAAUUCAGACCAACUACUGAAUGCUGACGGUUUAUGAUGAUUAAAAGGAGAGCAUGUAUUUUGAAUAC ((((.(((((((((----.............((((((........)))))).((...((((......))))...)).))))))))).))))...... ( -17.00, z-score = -2.47, R) >droSim1.chr3R 1782998 96 - 27517382 GAAACACAUGCUUUUGAAAAAAAAAACACAAAUUCAGACCAACUAAUUCC-GCUGACGGUCUAUGAUGAUUAAAAAAACAGCAUGUAUUUUGAAAAC ((((.((((((((((....))))............(((((..(.......-...)..)))))..................)))))).))))...... ( -15.10, z-score = -2.06, R) >droSec1.super_6 1856829 94 - 4358794 GAAACACAUGCUUUUG--AAAAAAAAUACAAACUUAGACCAACUAAUUCA-GCUGACGGUCUAUGAUAAUUAAAAGAACAGCAUGUCUUUUGAAAAC ((((.((((((((((.--...)))).......(.((((((..(.......-...)..)))))).)...............)))))).))))...... ( -16.40, z-score = -2.59, R) >consensus GAAACACAUGCUUUUG__AAAAAAAAAACAAAUUCAGACCAACUAAUUCA_GCUGACGGUCUAUGAUGAUUAAAAGAACAGCAUGUAUUUUGAAAAC ((((.((((((((((....))))............(((((..(...........)..)))))..................)))))).))))...... (-12.22 = -12.33 + 0.11)

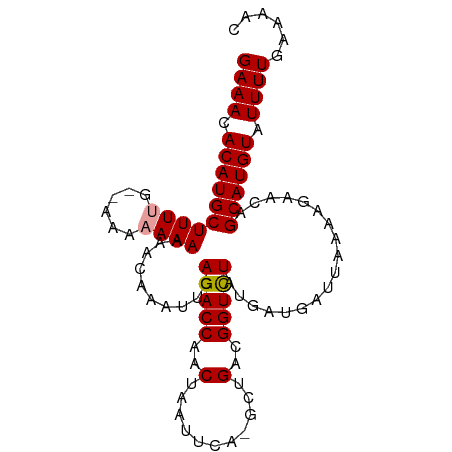
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:26 2011