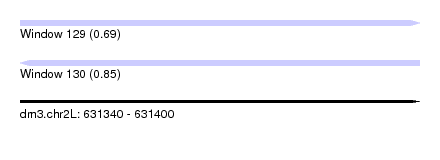
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 631,340 – 631,400 |
| Length | 60 |
| Max. P | 0.854142 |

| Location | 631,340 – 631,400 |
|---|---|
| Length | 60 |
| Sequences | 10 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 74.20 |
| Shannon entropy | 0.54766 |
| G+C content | 0.43836 |
| Mean single sequence MFE | -10.54 |
| Consensus MFE | -8.69 |
| Energy contribution | -8.21 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
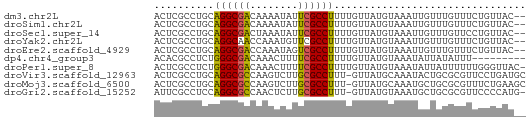
>dm3.chr2L 631340 60 + 23011544 ACUCGCCUGCAGGCGACAAAAUAUUCGCCUUUUGUUAUGUAAAUUGUUUGUUUCUGUUAC-- ..(((((....)))))(((((........)))))..........................-- ( -8.30, z-score = 0.01, R) >droSim1.chr2L 637817 60 + 22036055 ACUCGCCUGCAGGCGACAAAAUAUUCGCCUUUUGUUAUGUAAAUUGUUUGUUUCUGUUAC-- ..(((((....)))))(((((........)))))..........................-- ( -8.30, z-score = 0.01, R) >droSec1.super_14 613528 60 + 2068291 ACUCGCCUGCAGGCGACUAAAUAUUCGCCUUUUGUUAUGUAAAUUGUUUGUUCCUGUUAC-- ..........((((((........))))))..............................-- ( -8.10, z-score = -0.20, R) >droYak2.chr2L 621295 60 + 22324452 ACUCGCCUGCAGGCAACCAAAUGUUCGCCUUUUGUUAUGUAAAUUGUUUGUUUCUGUUAC-- ..........(((((((.....))).))))..............................-- ( -5.30, z-score = 1.37, R) >droEre2.scaffold_4929 689302 60 + 26641161 ACUCGCCUGCAGGCGACCAAAUAGUCGCCUUUUGUUAUGUAAAUUGUUUGUUUCUGUUAC-- ..........(((((((......)))))))..............................-- ( -11.30, z-score = -0.99, R) >dp4.chr4_group3 2593860 53 - 11692001 ACACGCCUCUGGGCGACAAACUUUUCGCCUUUUGUUAUGUAAAUAUUAUAUUU--------- (((.......((((((........)))))).......))).............--------- ( -9.14, z-score = -1.69, R) >droPer1.super_8 3683466 61 - 3966273 ACUCGCCUCUGGGCGACAAACUUUUCGCCUUUUGUUAUGUAAAUAUUAUUUUUUGGGUUAC- ..(((((....)))))..........((((...(..(((.......)))..)..))))...- ( -9.80, z-score = -0.80, R) >droVir3.scaffold_12963 19107327 61 + 20206255 ACUCGCCUGCAGGCGCCAAGUCUUGCGCCUUU-GUUAUGCAAAUACUGCGCGUUCCUGAUGC ...(((..((((((((........)))))(((-(.....))))...)))))).......... ( -14.80, z-score = 0.37, R) >droMoj3.scaffold_6500 8253243 61 - 32352404 ACUCGCCUGCAGGCGCCAAGUCUUGCGCCUUU-GUUAUGCAAAUGCUGCGCGUUUCUGAAGC ((.(((..((((((((........)))))(((-(.....))))))).))).))......... ( -17.60, z-score = 0.05, R) >droGri2.scaffold_15252 6992662 60 + 17193109 AUUCGCCUCCAGGCGCCAACUCUUGCGCCUUU-GUUAUGUAAAUGCUGCGCGUUCCCCAUG- ...(((....((((((........))))))..-.....(((.....)))))).........- ( -12.80, z-score = -0.92, R) >consensus ACUCGCCUGCAGGCGACAAAAUUUUCGCCUUUUGUUAUGUAAAUUGUUUGUUUCUGUUAC__ ..........((((((........))))))................................ ( -8.69 = -8.21 + -0.48)

| Location | 631,340 – 631,400 |
|---|---|
| Length | 60 |
| Sequences | 10 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 74.20 |
| Shannon entropy | 0.54766 |
| G+C content | 0.43836 |
| Mean single sequence MFE | -11.48 |
| Consensus MFE | -9.03 |
| Energy contribution | -8.72 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
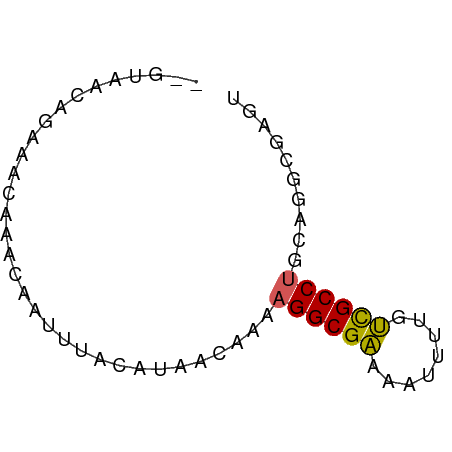
>dm3.chr2L 631340 60 - 23011544 --GUAACAGAAACAAACAAUUUACAUAACAAAAGGCGAAUAUUUUGUCGCCUGCAGGCGAGU --..........................(((((........)))))(((((....))))).. ( -9.50, z-score = -0.87, R) >droSim1.chr2L 637817 60 - 22036055 --GUAACAGAAACAAACAAUUUACAUAACAAAAGGCGAAUAUUUUGUCGCCUGCAGGCGAGU --..........................(((((........)))))(((((....))))).. ( -9.50, z-score = -0.87, R) >droSec1.super_14 613528 60 - 2068291 --GUAACAGGAACAAACAAUUUACAUAACAAAAGGCGAAUAUUUAGUCGCCUGCAGGCGAGU --((((...(......)...))))........((((((........)))))).......... ( -9.00, z-score = -0.82, R) >droYak2.chr2L 621295 60 - 22324452 --GUAACAGAAACAAACAAUUUACAUAACAAAAGGCGAACAUUUGGUUGCCUGCAGGCGAGU --..........................((((.(.....).)))).(((((....))))).. ( -7.40, z-score = 0.51, R) >droEre2.scaffold_4929 689302 60 - 26641161 --GUAACAGAAACAAACAAUUUACAUAACAAAAGGCGACUAUUUGGUCGCCUGCAGGCGAGU --..............................((((((((....)))))))).......... ( -13.30, z-score = -2.13, R) >dp4.chr4_group3 2593860 53 + 11692001 ---------AAAUAUAAUAUUUACAUAACAAAAGGCGAAAAGUUUGUCGCCCAGAGGCGUGU ---------.............((((..(....(((((........)))))....)..)))) ( -9.20, z-score = -1.25, R) >droPer1.super_8 3683466 61 + 3966273 -GUAACCCAAAAAAUAAUAUUUACAUAACAAAAGGCGAAAAGUUUGUCGCCCAGAGGCGAGU -...............................((((.....)))).(((((....))))).. ( -8.10, z-score = -0.82, R) >droVir3.scaffold_12963 19107327 61 - 20206255 GCAUCAGGAACGCGCAGUAUUUGCAUAAC-AAAGGCGCAAGACUUGGCGCCUGCAGGCGAGU ..........(((((((...)))).....-..((((((........))))))....)))... ( -17.30, z-score = -0.01, R) >droMoj3.scaffold_6500 8253243 61 + 32352404 GCUUCAGAAACGCGCAGCAUUUGCAUAAC-AAAGGCGCAAGACUUGGCGCCUGCAGGCGAGU (((.(........).)))((((((.....-..((((((........))))))....)))))) ( -17.70, z-score = 0.24, R) >droGri2.scaffold_15252 6992662 60 - 17193109 -CAUGGGGAACGCGCAGCAUUUACAUAAC-AAAGGCGCAAGAGUUGGCGCCUGGAGGCGAAU -...........(((..(...........-..((((((........)))))).)..)))... ( -13.84, z-score = -0.11, R) >consensus __GUAACAGAAACAAACAAUUUACAUAACAAAAGGCGAAAAUUUUGUCGCCUGCAGGCGAGU ................................((((((........)))))).......... ( -9.03 = -8.72 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:27 2011