| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,721,602 – 1,721,652 |
| Length | 50 |
| Max. P | 0.947273 |

| Location | 1,721,602 – 1,721,652 |
|---|---|
| Length | 50 |
| Sequences | 11 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 81.08 |
| Shannon entropy | 0.40134 |
| G+C content | 0.43723 |
| Mean single sequence MFE | -14.01 |
| Consensus MFE | -9.52 |
| Energy contribution | -10.75 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
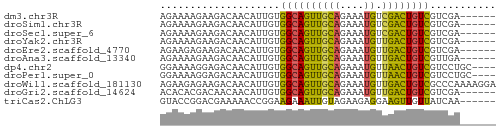
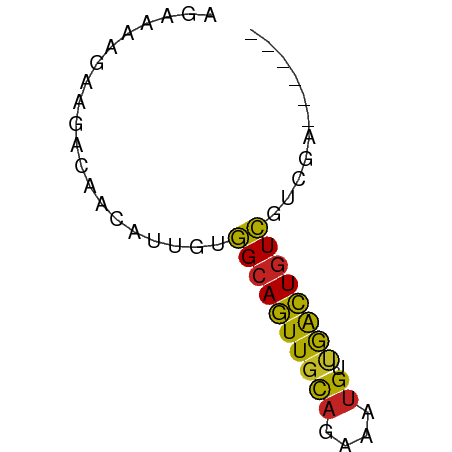
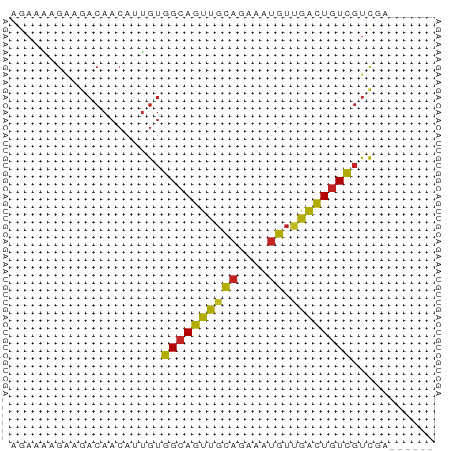
>dm3.chr3R 1721602 50 + 27905053 AGAAAAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUCGACUGUCGUCGA------ .........(((.((...))((((((((((....)).)))))))))))..------ ( -14.90, z-score = -2.20, R) >droSim1.chr3R 1755017 50 + 27517382 AGAAAAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUCGACUGUCGUCGA------ .........(((.((...))((((((((((....)).)))))))))))..------ ( -14.90, z-score = -2.20, R) >droSec1.super_6 1829554 50 + 4358794 AGAAAAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUCGACUGUCGUCGA------ .........(((.((...))((((((((((....)).)))))))))))..------ ( -14.90, z-score = -2.20, R) >droYak2.chr3R 17666522 50 + 28832112 AGAAAAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUUGACUGUCGUCGA------ .........(((.((...))((((((((((....)).)))))))))))..------ ( -12.30, z-score = -1.34, R) >droEre2.scaffold_4770 1989096 50 + 17746568 AGAAGAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUUGACUGUCGUCGA------ .((.((..((.(((((((...((....))...))))))).)).)).))..------ ( -13.00, z-score = -1.45, R) >droAna3.scaffold_13340 5314130 50 - 23697760 AGAAAAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUUGACUGUCGUUGA------ ...........((((.....((((((((((....)).)))))))))))).------ ( -12.50, z-score = -1.71, R) >dp4.chr2 14736534 52 - 30794189 GGAAAAGGAGACAACAUUGUGGCAGUUGCAGAAAUGUUAACUGUCGUCCUGC---- (((...(....)........((((((((((....)).)))))))).)))...---- ( -16.80, z-score = -2.77, R) >droPer1.super_0 4352577 52 + 11822988 GGAAAAGGAGACAACAUUGUGGCAGUUGCAGAAAUGUUAACUGUCGUCCUGC---- (((...(....)........((((((((((....)).)))))))).)))...---- ( -16.80, z-score = -2.77, R) >droWil1.scaffold_181130 5083327 56 - 16660200 AGAAGAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUUGACUGUCGCCCAAAAGGA ..................((((((((((((....)).))))))))))((....)). ( -15.00, z-score = -2.27, R) >droGri2.scaffold_14624 1089449 50 - 4233967 ACACACGACAACAACAUUGUGGCAGUUGCAGAAAUGUUGACUGUCGUCGA------ .....((((.(((....)))((((((((((....)).)))))))))))).------ ( -16.90, z-score = -1.91, R) >triCas2.ChLG3 29421190 50 + 32080666 GUACCGGACGAAAAACCGGAAGAAAUUGUAGAAGAGGAAGUUGUUAUCAA------ ...((((........))))...............................------ ( -6.10, z-score = 0.05, R) >consensus AGAAAAGAAGACAACAUUGUGGCAGUUGCAGAAAUGUUGACUGUCGUCGA______ ....................((((((((((....)).))))))))........... ( -9.52 = -10.75 + 1.23)
| Location | 1,721,602 – 1,721,652 |
|---|---|
| Length | 50 |
| Sequences | 11 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Shannon entropy | 0.40134 |
| G+C content | 0.43723 |
| Mean single sequence MFE | -10.61 |
| Consensus MFE | -6.03 |
| Energy contribution | -6.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1721602 50 - 27905053 ------UCGACGACAGUCGACAUUUCUGCAACUGCCACAAUGUUGUCUUCUUUUCU ------.....((.((.(((((((...((....))...))))))).))))...... ( -9.70, z-score = -1.39, R) >droSim1.chr3R 1755017 50 - 27517382 ------UCGACGACAGUCGACAUUUCUGCAACUGCCACAAUGUUGUCUUCUUUUCU ------.....((.((.(((((((...((....))...))))))).))))...... ( -9.70, z-score = -1.39, R) >droSec1.super_6 1829554 50 - 4358794 ------UCGACGACAGUCGACAUUUCUGCAACUGCCACAAUGUUGUCUUCUUUUCU ------.....((.((.(((((((...((....))...))))))).))))...... ( -9.70, z-score = -1.39, R) >droYak2.chr3R 17666522 50 - 28832112 ------UCGACGACAGUCAACAUUUCUGCAACUGCCACAAUGUUGUCUUCUUUUCU ------.....((.((.(((((((...((....))...))))))).))))...... ( -10.00, z-score = -2.04, R) >droEre2.scaffold_4770 1989096 50 - 17746568 ------UCGACGACAGUCAACAUUUCUGCAACUGCCACAAUGUUGUCUUCUCUUCU ------..((.((.((.(((((((...((....))...))))))).))..)).)). ( -10.80, z-score = -2.44, R) >droAna3.scaffold_13340 5314130 50 + 23697760 ------UCAACGACAGUCAACAUUUCUGCAACUGCCACAAUGUUGUCUUCUUUUCU ------.....((.((.(((((((...((....))...))))))).))))...... ( -10.00, z-score = -2.63, R) >dp4.chr2 14736534 52 + 30794189 ----GCAGGACGACAGUUAACAUUUCUGCAACUGCCACAAUGUUGUCUCCUUUUCC ----..((((.((((....(((((...((....))...)))))))))))))..... ( -11.30, z-score = -1.72, R) >droPer1.super_0 4352577 52 - 11822988 ----GCAGGACGACAGUUAACAUUUCUGCAACUGCCACAAUGUUGUCUCCUUUUCC ----..((((.((((....(((((...((....))...)))))))))))))..... ( -11.30, z-score = -1.72, R) >droWil1.scaffold_181130 5083327 56 + 16660200 UCCUUUUGGGCGACAGUCAACAUUUCUGCAACUGCCACAAUGUUGUCUUCUCUUCU (((....))).((.((.(((((((...((....))...))))))).))))...... ( -11.20, z-score = -1.62, R) >droGri2.scaffold_14624 1089449 50 + 4233967 ------UCGACGACAGUCAACAUUUCUGCAACUGCCACAAUGUUGUUGUCGUGUGU ------...(((((((.(((((((...((....))...)))))))))))))).... ( -17.40, z-score = -2.37, R) >triCas2.ChLG3 29421190 50 - 32080666 ------UUGAUAACAACUUCCUCUUCUACAAUUUCUUCCGGUUUUUCGUCCGGUAC ------...............................((((........))))... ( -5.60, z-score = -0.63, R) >consensus ______UCGACGACAGUCAACAUUUCUGCAACUGCCACAAUGUUGUCUUCUUUUCU ..............((.(((((((...((....))...))))))).))........ ( -6.03 = -6.24 + 0.20)

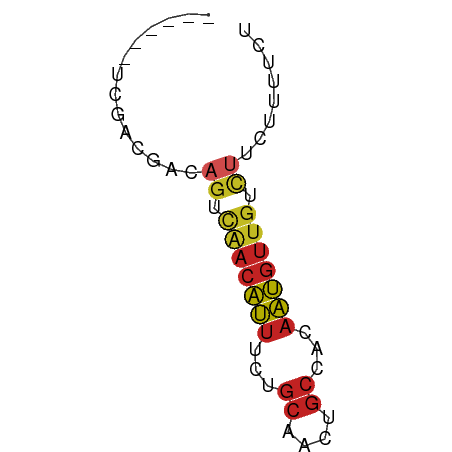
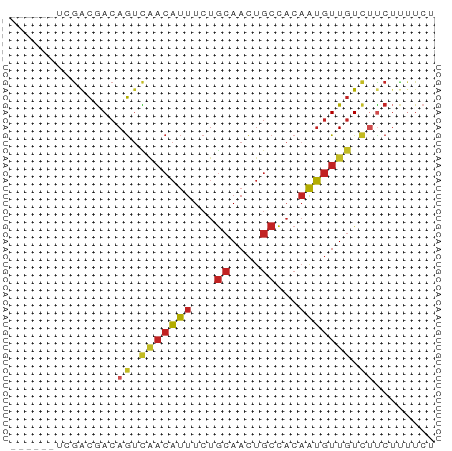
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:23 2011