
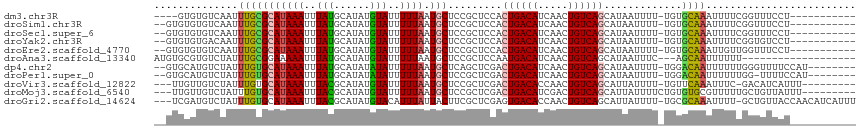
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,709,951 – 1,710,052 |
| Length | 101 |
| Max. P | 0.628355 |

| Location | 1,709,951 – 1,710,052 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Shannon entropy | 0.37802 |
| G+C content | 0.34683 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.25 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
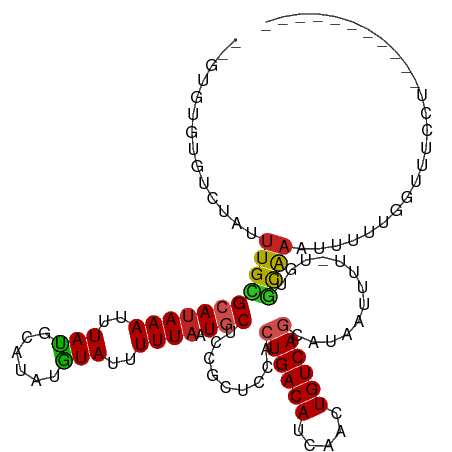
>dm3.chr3R 1709951 101 + 27905053 ----GUGUGUCAAUUUGCGCAUAAAUUUAUGCAUAUGUAUUUUUAAUGCUCCGCUCCACUGACAUCAACUGUCAGCAUAAUUUU-UGUGCAAAUUUUCGGUUUCCU----------- ----...((..((((((((((.(((.((((((....((((.....))))..........(((((.....))))))))))).)))-))))))))))..)).......----------- ( -24.30, z-score = -2.88, R) >droSim1.chr3R 1743407 103 + 27517382 --GUGUGUGUCAAUUUGCGCAUAAAUUUAUGCAUAUGUAUUUUUAAUGCUCCGCUCCACUGACAUCAACUGUCAGCAUAAUUUU-UGUGCAAAUUUUCGGUUUCCU----------- --.....((..((((((((((.(((.((((((....((((.....))))..........(((((.....))))))))))).)))-))))))))))..)).......----------- ( -24.30, z-score = -2.45, R) >droSec1.super_6 1817845 103 + 4358794 --GUGUGUGUCAAUUUGCGCAUAAAUUUAUGCAUAUGUAUUUUUAAUGCUCCGCUCCACUGACAUCAACUGUCAGCAUAAUUUU-UGUGCAAAUUUUCGGUUUCCU----------- --.....((..((((((((((.(((.((((((....((((.....))))..........(((((.....))))))))))).)))-))))))))))..)).......----------- ( -24.30, z-score = -2.45, R) >droYak2.chr3R 17655154 103 + 28832112 --GUGUGUGACAAUUUGCGCAUAAAUUUAUGCAUAUGUAUUUUUAAUGCUCCGCUCCACUGACAUCAACUGUCAGCAUAAUUUU-UGUGCAAAUUUUCGGUGUCCU----------- --.....(((.((((((((((.(((.((((((....((((.....))))..........(((((.....))))))))))).)))-)))))))))).))).......----------- ( -26.10, z-score = -2.36, R) >droEre2.scaffold_4770 1978172 103 + 17746568 --GUGUGUGUCAAUUUGCGCAUAAAUUUAUGCAUAUGUAUUUUUAAUGCUCCGCUCCACUGACAUCAACUGUCAGCAUAAUUUU-UGUGCAAAUUGUUGGUUUCCU----------- --........(((((((((((.(((.((((((....((((.....))))..........(((((.....))))))))))).)))-)))))))))))..........----------- ( -26.30, z-score = -2.57, R) >droAna3.scaffold_13340 5302939 95 - 23697760 AUGUGCGUGUCUAUUUGCGGAAAAAUUUAUGCAUAUGUAUUUUUAAUGCUCCGCUCCAAUGACAUCAACUGUCAGCAUAAUUUC---AGCAAUUUUUU------------------- .(((((..........(((((...(((.((((....))))....)))..))))).....(((((.....)))))))))).....---...........------------------- ( -19.50, z-score = -1.12, R) >dp4.chr2 14727108 106 - 30794189 --GUGCAUGUCUAUUUGUGCAUAAAUUUAUGCAUAUAUAUUUUUAAUGCUCAGCUCGACUGACAUCAACUGUCAGCAUAAUUUU-UGGACAAUUUUUUGGGUUUUCCAU-------- --(.((((...(((.((((((((....)))))))).)))......)))).).....(.((((((.....)))))))........-((((.((((.....)))).)))).-------- ( -23.80, z-score = -1.82, R) >droPer1.super_0 4342924 105 + 11822988 --GUGCAUGUCUAUUUGUGCAUAAAUUUAUGCAUAUAUAUUUUUAAUGCUCCGCUCGACUGACAUCAACUGUCAGCAUAAUUUU-UGGACAAUUUUUUGG-UUUUCCAU-------- --..((((...(((.((((((((....)))))))).)))......)))).......(.((((((.....)))))))........-((((.((((....))-)).)))).-------- ( -23.20, z-score = -2.12, R) >droVir3.scaffold_12822 1881222 103 - 4096053 ---UUGUUGUCUAUUUGUGCAUAAAUUUACGCAUAUGUAUUUUUAAUGCUCCGCUCGACUGACACCAACUGUCAGCAUUAUUUU-UGUUCAAAUUUC-GACAUCAUUU--------- ---.(((((...(((((.(((.((((....((....((((.....))))...))..(.((((((.....)))))))...)))).-))).)))))..)-))))......--------- ( -20.10, z-score = -2.12, R) >droMoj3.scaffold_6540 10979185 105 + 34148556 ---UUGUUGUCUAUUUGUGCAUAAAUUUACGCAUAUGUAUUUUUAAUGCUCCGCUCGACUGACAUCGACUGUCAGCAUUAUUUUCUGUGUGCGUUUUUGCUGUUAUUU--------- ---...............(((.......((((((((.......(((((((.((.((((......)))).))..)))))))......))))))))...)))........--------- ( -21.92, z-score = -1.36, R) >droGri2.scaffold_14624 1079125 112 - 4233967 ---UCGAUGUCUAUUUGUGCAUAAAUUUACGCAUAUGUACAUUUAUUACUUCGCUCGAGUGACACCAACUGUCAGCAUUAUUUU-UGCGCAAAUUUU-GCUGUUACCAACAUCAUUU ---..(((((.((..((((((((..........))))))))..))...........(.((((((..........(((.......-)))(((.....)-))))))))).))))).... ( -22.20, z-score = -1.40, R) >consensus __GUGUGUGUCUAUUUGCGCAUAAAUUUAUGCAUAUGUAUUUUUAAUGCUCCGCUCCACUGACAUCAACUGUCAGCAUAAUUUU_UGUGCAAAUUUUUGGUUUCCU___________ ..............(((((((((((..(((......)))..)))).))).........((((((.....)))))).............))))......................... (-11.46 = -11.25 + -0.20)
| Location | 1,709,951 – 1,710,052 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.66 |
| Shannon entropy | 0.37802 |
| G+C content | 0.34683 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -12.78 |
| Energy contribution | -13.42 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
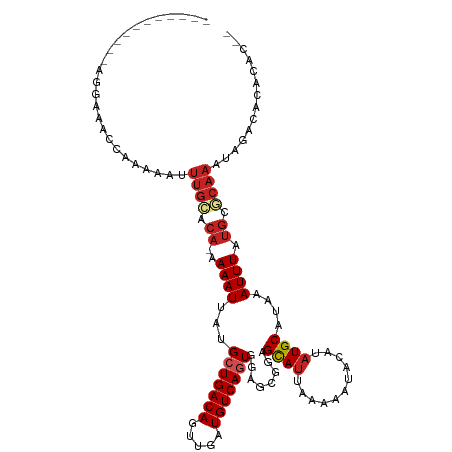
>dm3.chr3R 1709951 101 - 27905053 -----------AGGAAACCGAAAAUUUGCACA-AAAAUUAUGCUGACAGUUGAUGUCAGUGGAGCGGAGCAUUAAAAAUACAUAUGCAUAAAUUUAUGCGCAAAUUGACACAC---- -----------.(....).(..(((((((.((-.(((((..(((((((.....)))))))........((((...........))))...))))).)).)))))))..)....---- ( -23.50, z-score = -2.29, R) >droSim1.chr3R 1743407 103 - 27517382 -----------AGGAAACCGAAAAUUUGCACA-AAAAUUAUGCUGACAGUUGAUGUCAGUGGAGCGGAGCAUUAAAAAUACAUAUGCAUAAAUUUAUGCGCAAAUUGACACACAC-- -----------.(....).(..(((((((.((-.(((((..(((((((.....)))))))........((((...........))))...))))).)).)))))))..)......-- ( -23.50, z-score = -2.20, R) >droSec1.super_6 1817845 103 - 4358794 -----------AGGAAACCGAAAAUUUGCACA-AAAAUUAUGCUGACAGUUGAUGUCAGUGGAGCGGAGCAUUAAAAAUACAUAUGCAUAAAUUUAUGCGCAAAUUGACACACAC-- -----------.(....).(..(((((((.((-.(((((..(((((((.....)))))))........((((...........))))...))))).)).)))))))..)......-- ( -23.50, z-score = -2.20, R) >droYak2.chr3R 17655154 103 - 28832112 -----------AGGACACCGAAAAUUUGCACA-AAAAUUAUGCUGACAGUUGAUGUCAGUGGAGCGGAGCAUUAAAAAUACAUAUGCAUAAAUUUAUGCGCAAAUUGUCACACAC-- -----------........((.(((((((.((-.(((((..(((((((.....)))))))........((((...........))))...))))).)).))))))).))......-- ( -24.50, z-score = -1.91, R) >droEre2.scaffold_4770 1978172 103 - 17746568 -----------AGGAAACCAACAAUUUGCACA-AAAAUUAUGCUGACAGUUGAUGUCAGUGGAGCGGAGCAUUAAAAAUACAUAUGCAUAAAUUUAUGCGCAAAUUGACACACAC-- -----------.(....)...((((((((.((-.(((((..(((((((.....)))))))........((((...........))))...))))).)).))))))))........-- ( -26.30, z-score = -3.13, R) >droAna3.scaffold_13340 5302939 95 + 23697760 -------------------AAAAAAUUGCU---GAAAUUAUGCUGACAGUUGAUGUCAUUGGAGCGGAGCAUUAAAAAUACAUAUGCAUAAAUUUUUCCGCAAAUAGACACGCACAU -------------------.......((((---(.........(((((.....))))).....(((((((((...........)))).........))))).......)).)))... ( -16.50, z-score = -0.06, R) >dp4.chr2 14727108 106 + 30794189 --------AUGGAAAACCCAAAAAAUUGUCCA-AAAAUUAUGCUGACAGUUGAUGUCAGUCGAGCUGAGCAUUAAAAAUAUAUAUGCAUAAAUUUAUGCACAAAUAGACAUGCAC-- --------.(((.....)))......((((..-......(((((..((((((((....))).))))))))))............((((((....))))))......)))).....-- ( -20.10, z-score = -1.34, R) >droPer1.super_0 4342924 105 - 11822988 --------AUGGAAAA-CCAAAAAAUUGUCCA-AAAAUUAUGCUGACAGUUGAUGUCAGUCGAGCGGAGCAUUAAAAAUAUAUAUGCAUAAAUUUAUGCACAAAUAGACAUGCAC-- --------.(((....-)))........(((.-........(((((((.....))))))).....)))((((......(((.(.((((((....)))))).).)))...))))..-- ( -19.04, z-score = -0.84, R) >droVir3.scaffold_12822 1881222 103 + 4096053 ---------AAAUGAUGUC-GAAAUUUGAACA-AAAAUAAUGCUGACAGUUGGUGUCAGUCGAGCGGAGCAUUAAAAAUACAUAUGCGUAAAUUUAUGCACAAAUAGACAACAA--- ---------...((.((((-...(((((....-....(((((((((((.....)))).(.....)..)))))))..........((((((....))))))))))).)))).)).--- ( -21.60, z-score = -1.47, R) >droMoj3.scaffold_6540 10979185 105 - 34148556 ---------AAAUAACAGCAAAAACGCACACAGAAAAUAAUGCUGACAGUCGAUGUCAGUCGAGCGGAGCAUUAAAAAUACAUAUGCGUAAAUUUAUGCACAAAUAGACAACAA--- ---------........(((((((((((.........(((((((..(..(((((....)))))..).)))))))..........)))))...))).)))...............--- ( -18.51, z-score = -1.29, R) >droGri2.scaffold_14624 1079125 112 + 4233967 AAAUGAUGUUGGUAACAGC-AAAAUUUGCGCA-AAAAUAAUGCUGACAGUUGGUGUCACUCGAGCGAAGUAAUAAAUGUACAUAUGCGUAAAUUUAUGCACAAAUAGACAUCGA--- ...(((((((.......((-((((((((((((-.......((((((((.....)))).....))))..(((.......)))...))))))))))).))).......))))))).--- ( -27.44, z-score = -1.52, R) >consensus ___________AGGAAACCAAAAAUUUGCACA_AAAAUUAUGCUGACAGUUGAUGUCAGUGGAGCGGAGCAUUAAAAAUACAUAUGCAUAAAUUUAUGCGCAAAUAGACACACAC__ .........................((((.((.(((.....(((((((.....)))))))........((((...........)))).....))).)).)))).............. (-12.78 = -13.42 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:20 2011