| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,642,490 – 1,642,582 |
| Length | 92 |
| Max. P | 0.994455 |

| Location | 1,642,490 – 1,642,582 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 65.70 |
| Shannon entropy | 0.57962 |
| G+C content | 0.60308 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -11.08 |
| Energy contribution | -11.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
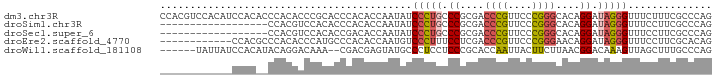
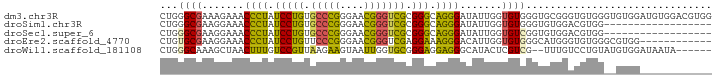
>dm3.chr3R 1642490 92 + 27905053 CCACGUCCACAUCCACACCCACACCCGCACCCACACCAAUAUCCCUGCCCGCGACCCGUUCCCGGGCACAGGAUAGGGUUUCUUUCGCCCAG .......................................(((((.((((((.((.....)).))))))..)))))((((.......)))).. ( -21.90, z-score = -2.40, R) >droSim1.chr3R 1675505 74 + 27517382 ------------------CCACGUCCACACCCACACCAAUAUCCCUGCCCGCGACCCGUUCCCGGGCACAGGAUAGGGUUUCCUUCGCCCAG ------------------.....................(((((.((((((.((.....)).))))))..)))))((((.......)))).. ( -21.90, z-score = -2.77, R) >droSec1.super_6 1750634 74 + 4358794 ------------------CCACGUCCACACCGACACCAAUAUCCCUGCCCGCGACCCGUUCCCGGGCACAGGAUAGGGUUUCCUUCGCCCAG ------------------....(((......))).....(((((.((((((.((.....)).))))))..)))))((((.......)))).. ( -24.00, z-score = -3.02, R) >droEre2.scaffold_4770 1909637 80 + 17746568 ------------CCACGCCCACACCCAUGCCCACACCAAUGUCCCUUUCCUCGACCCGUUCCCGGGAACAGGAUAGGGUUUCCUUCGCACAG ------------...............(((..(((....)))((((.((((...((((....))))...)))).))))........)))... ( -18.40, z-score = -1.08, R) >droWil1.scaffold_181108 3883067 84 - 4707319 ------UAUUAUCCACAUACAGGACAAA--CGACGAGUAUGCCCUCCUCCCGCACCAAUUACUUCUUAACGGACAAAGUUAGCUUUGCCCAG ------..............(((.((.(--(.....)).)).))).........................((.(((((....))))).)).. ( -11.60, z-score = -0.80, R) >consensus ____________CCAC__CCACGCCCACACCCACACCAAUAUCCCUGCCCGCGACCCGUUCCCGGGCACAGGAUAGGGUUUCCUUCGCCCAG ..........................................(((((.((....((((....))))....)).))))).............. (-11.08 = -11.32 + 0.24)
| Location | 1,642,490 – 1,642,582 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 65.70 |
| Shannon entropy | 0.57962 |
| G+C content | 0.60308 |
| Mean single sequence MFE | -29.80 |
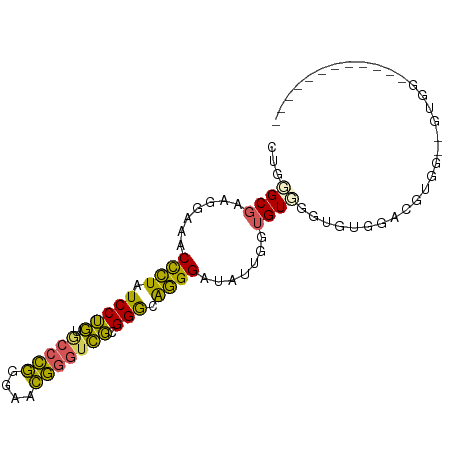
| Consensus MFE | -19.54 |
| Energy contribution | -18.30 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
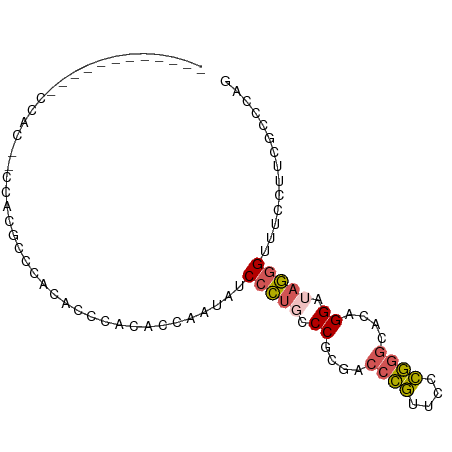
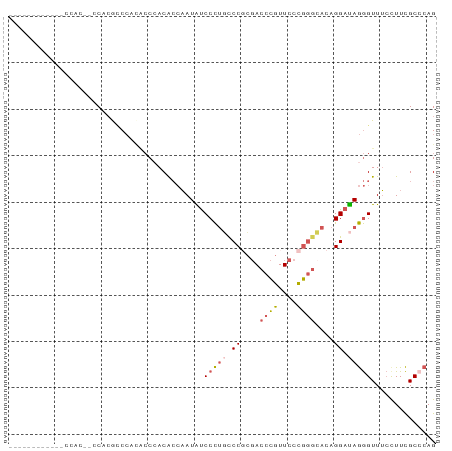
>dm3.chr3R 1642490 92 - 27905053 CUGGGCGAAAGAAACCCUAUCCUGUGCCCGGGAACGGGUCGCGGGCAGGGAUAUUGGUGUGGGUGCGGGUGUGGGUGUGGAUGUGGACGUGG (((.((........((((..((((((((((....)))).)))))).))))(((((.((......)).)))))..)).)))............ ( -31.50, z-score = -0.56, R) >droSim1.chr3R 1675505 74 - 27517382 CUGGGCGAAGGAAACCCUAUCCUGUGCCCGGGAACGGGUCGCGGGCAGGGAUAUUGGUGUGGGUGUGGACGUGG------------------ (((.((.((.....((((..((((((((((....)))).)))))).))))...)).)).)))............------------------ ( -28.30, z-score = -1.09, R) >droSec1.super_6 1750634 74 - 4358794 CUGGGCGAAGGAAACCCUAUCCUGUGCCCGGGAACGGGUCGCGGGCAGGGAUAUUGGUGUCGGUGUGGACGUGG------------------ ....(((.......((((..((((((((((....)))).)))))).))))((((((....))))))...)))..------------------ ( -29.30, z-score = -1.13, R) >droEre2.scaffold_4770 1909637 80 - 17746568 CUGUGCGAAGGAAACCCUAUCCUGUUCCCGGGAACGGGUCGAGGAAAGGGACAUUGGUGUGGGCAUGGGUGUGGGCGUGG------------ (..(((........((((.((((...((((....))))...)))).))))(((((.(((....))).)))))..)))..)------------ ( -34.00, z-score = -2.97, R) >droWil1.scaffold_181108 3883067 84 + 4707319 CUGGGCAAAGCUAACUUUGUCCGUUAAGAAGUAAUUGGUGCGGGAGGAGGGCAUACUCGUCG--UUUGUCCUGUAUGUGGAUAAUA------ ..((((((((....))))))))...............((((((((.((((((......))).--))).))))))))..........------ ( -25.90, z-score = -2.16, R) >consensus CUGGGCGAAGGAAACCCUAUCCUGUGCCCGGGAACGGGUCGCGGGCAGGGAUAUUGGUGUGGGUGUGGACGUGG__GUGG____________ ...((((.......((((.(((((.(((((....))))))).))).)))).......))))............................... (-19.54 = -18.30 + -1.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:14 2011