
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,633,024 – 1,633,170 |
| Length | 146 |
| Max. P | 0.698237 |

| Location | 1,633,024 – 1,633,170 |
|---|---|
| Length | 146 |
| Sequences | 9 |
| Columns | 152 |
| Reading direction | forward |
| Mean pairwise identity | 70.55 |
| Shannon entropy | 0.61091 |
| G+C content | 0.55498 |
| Mean single sequence MFE | -40.64 |
| Consensus MFE | -21.02 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.698237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1633024 146 + 27905053 CUGCCGCCGCCGCCUCAAAUCGAGCUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUCCGUCGUCCGCUGGUGCGCAUUAGACGUGAGUAA-AUCCUCCUAAAC-AUCCGUA-AU-CCACC--CGGAUACUCGUUUCGCUCAAGUCCCCU .(((((((((((.(((.....))).))))))))).)).((.(((((.(((((.((.....)))))))....)))))))....(((.(((((..-..........((-....)).-((-((...--.)))).........))))).))).... ( -47.90, z-score = -2.38, R) >droSim1.chr3R 1666053 146 + 27517382 CUGCCGCCGCCGCCUCAAAUCGAGCUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUCCGGCGUCCGCUGGUGCGCAUUAGACGUGAGUAA-AUCCUUCUAAAC-AUUCAUA-AU-CCACC--CGGAUACUCGUUUCGCUCAAGUCCCCU .(((((((((((.(((.....))).))))))))).))....((.....(((((.((((.(((((....))))).)))).(((((.(.((....-.))).)))))..-.))))).-((-((...--.)))).........))........... ( -48.60, z-score = -2.10, R) >droSec1.super_6 1741305 146 + 4358794 CUGCCGCCGCCGCCUCAAAUCGAGCUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUCCGGCGUCCGCUGGUGCGCAUUAGACGUGAGUAA-AUCCUUCUAAAC-AUCCAUA-AU-CCACC--CGGAUACUCGUUUCGCUCAAGUCCCCU .(((((((((((.(((.....))).))))))))).))....((....((((...((((.(((((....))))).)))).(((((.(.((....-.))).))))).)-)))....-((-((...--.)))).........))........... ( -49.20, z-score = -2.31, R) >droEre2.scaffold_4770 1899942 146 + 17746568 CUGCCGCCGCCACCGCAAAUCGAGCUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUUCGCCGUCCGCUGGUGCGCAUUAGACGUGAGUAA-AUCCCCCUAAAC-AACUUUA-AU-CCAUC--CGGAUACUCGUUUCGCCUAAGUAGCCU ((((.(((((((((((.......)).)))))))).)..((((((((.(.((.(((.((...))))).))).)))))).....((..((((((.-.......((((.-...))))-.(-((...--.)))))))))..))))....))))... ( -43.90, z-score = -1.07, R) >droAna3.scaffold_13340 5228783 113 - 23697760 CUGCCGCCGCCCCCCCAAAUCGAGCUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUCCGGCGCCCGCUAGUGCGCAUUAGACGUAAGUAA-AGACUCUUAAUC-CUUUCUA-A------------------------------------ .((((((((((..(.(.....).)..)))))))).))........((((((..((..((..(((....)))))..))))))))((....))..-............-.......-.------------------------------------ ( -29.30, z-score = 0.33, R) >dp4.chr2 18439429 131 - 30794189 CUGCCGCCGCCGCCACAAAUCGAACUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUGCGACGCCCAUUGGUGCGCAUUAGACGUGAGUAU-AUAUAGCCAAUCCGUUGAUA-AUUCCGUG--CAAAUCCCCU----------------- .(((.((((((((((..........))))))))).)..((.(((((.((((..(((......)))..))))))))))).....(((.((((.(-((.((((......)))))))-))))))))--))........----------------- ( -38.50, z-score = -0.62, R) >droPer1.super_0 8114399 131 + 11822988 CUGCCGCCGCCGCCACAAAUCGAGCUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUGCGACGCCCAUUGGUGCGCAUUAGACGUGAGUAU-AUAUAGCCAAUCCGUUGAUA-AUUCCGUG--CAAAUCCCCU----------------- .(((((((((((((.......).)).)))))))).))....((((...((.((((.((((((.((((....))))))).....((....))..-....))).....)))).)).-.....)))--).........----------------- ( -39.30, z-score = -0.45, R) >droMoj3.scaffold_6540 7950420 140 - 34148556 UUGCCGCCGCCGCCGCAAAUCGAACUGGUCGUGGAUAAGCCCCAGCUGAUGAAUGUAUUGCGUCGUCCACUGGUGCGAAUAAGACGUAAGUAUUAUGAUCCCACAGCAUGCCCAUAUAUUAUC--CUUAAGCGCACGACGCU---------- ((((.((....)).)))).........(((((((((.(((....)))((((.........))))))))))..(((((..((((((....))..((((...............)))).......--))))..))))))))...---------- ( -33.66, z-score = 0.90, R) >anoGam1.chr2R 51227219 139 + 62725911 CUUCCCCCACCGCCACAAAUUGAGCUGGUUGUCGAUAAACCACAUCUGAUGGGCGUUUUACGAAGACCUAUUGUACGGAUACGACGUGAGUUG---CGCCUAAUAGGGUCCCCAGAUACCAUCGCUUGGUCCUACUGAUGCU---------- ((((......((((.((....((..(((((.......)))))..))...))))))......)))).(((((((...((...((((....))))---..))))))))).....(((..((((.....))))....))).....---------- ( -35.40, z-score = 0.08, R) >consensus CUGCCGCCGCCGCCACAAAUCGAGCUGGUGGUGGACAAGCCGCACCUGAUGAACGUGCUCCGACGUCCGCUGGUGCGCAUUAGACGUGAGUAA_AUCCUCCUAAAC_AUCCAUA_AU_CCAUC__CGGAUACUCGUUUCGCU__________ ...(((((((((.(.(.....).).)))))))))....(((((..((((((..((((((............))))))))))))..))).))............................................................. (-21.02 = -22.00 + 0.98)
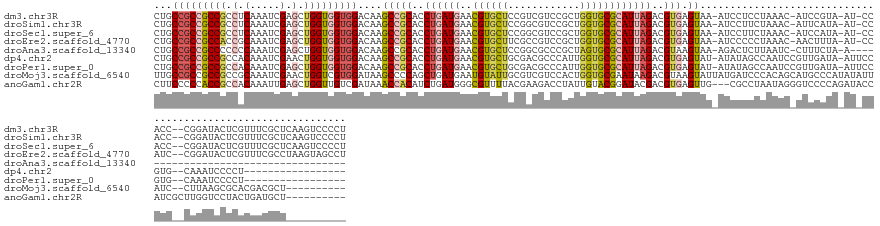


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:12 2011