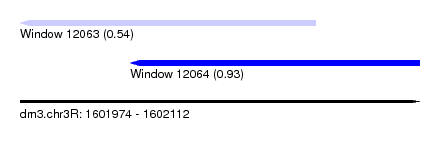
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,601,974 – 1,602,112 |
| Length | 138 |
| Max. P | 0.931933 |

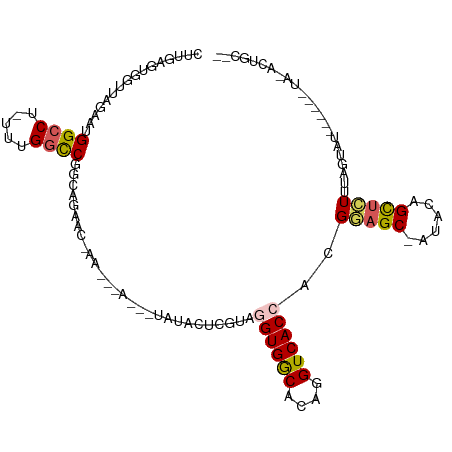
| Location | 1,601,974 – 1,602,076 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.12 |
| Shannon entropy | 0.55236 |
| G+C content | 0.44579 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -13.98 |
| Energy contribution | -16.73 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1601974 102 - 27905053 AAACUAAGGCU--AUACUCGUAGGUGGCACAGGUCACCACGGAGCAUACAGCUCUUUAGU--------AUUAAA----GAAACCUAGAUUGGAUGAUCCUGAUCCUUAUAGGGCAC ..((((((...--.....(((.((((((....)))))))))((((.....))))))))))--------......----....((((....((((.......))))...)))).... ( -28.60, z-score = -1.72, R) >droSim1.chr3R 1633133 111 - 27517382 ----UAAGACUAUAUACUCGUAGGUGGCACAGGUCACCACGGAGCGAACAGCUCUUUAGUAUUAAAUAACUGCAU-CUGCAACCUAGUGGGCAUGAUCCUGAUCCAUAUAGGGCAC ----(((.((((......(((.((((((....)))))))))((((.....))))..)))).)))......(((..-..))).(((((((((((......)).))))).)))).... ( -30.90, z-score = -1.30, R) >droSec1.super_6 1710423 99 - 4358794 ----UAAGACUGUAUACUCGUAGGUGGCACAGGUCACCACGGAGCG-----CUCUUUAGU-------AACUGCAU-CUGAAACCUAGUGAGGAUGAUCUUGAUCCUUAUAGGGCAC ----..(((.(((((((((((.((((((....)))))))))((...-----.))...)))-------)..)))))-))....(((.((((((((.......)))))))).)))... ( -27.80, z-score = -0.68, R) >droYak2.chr3R 17555787 103 - 28832112 ------------CAAAGGUAUAAGUGACACAGGUCACGCCGAAGCUAUAUAGUAUUAAAGAACUCCAAGUAACUUGCGGAAACCUAUUGAGAAUGAUCCUGAUCCUUGUAGGACA- ------------(((((((....(((((....))))).((((((.(((..(((........)))....))).))).)))..)))).))).......(((((.......)))))..- ( -19.80, z-score = -0.14, R) >consensus ____UAAGACU_UAUACUCGUAGGUGGCACAGGUCACCACGGAGCGAACAGCUCUUUAGU_______AACUACAU_CUGAAACCUAGUGAGGAUGAUCCUGAUCCUUAUAGGGCAC ..................(((.((((((....)))))))))((((.....))))............................((((.(((((((.......))))))))))).... (-13.98 = -16.73 + 2.75)

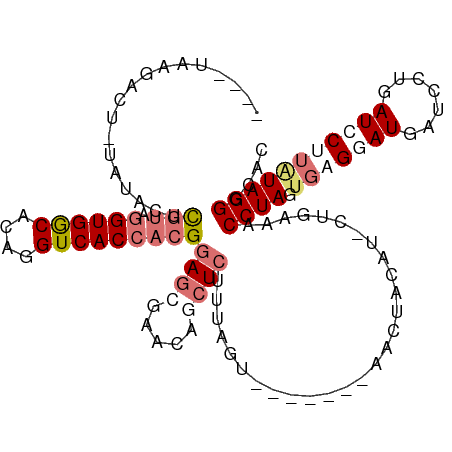
| Location | 1,602,012 – 1,602,112 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 64.43 |
| Shannon entropy | 0.62404 |
| G+C content | 0.46701 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.931933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1602012 100 - 27905053 CUUCAGUAGUUGGAAUGGCCU-UUUGGCCGGCAGAACAAACUAAGGCUAUACUCGUAGGUGGCACAGGUCACCACGGAGC-AUACAGCUCUUUAGUAU------UA-AA----- ........(((....(((((.-...)))))....)))..((((((........(((.((((((....)))))))))((((-.....))))))))))..------..-..----- ( -30.30, z-score = -1.77, R) >droSim1.chr3R 1633173 107 - 27517382 CUUGAGUGGUUAGAAUGGCCU-UUUGGCCGGCAGAACUAA--GACUAUAUACUCGUAGGUGGCACAGGUCACCACGGAGC-GAACAGCUCUUUAGUAU--UAAAUA-ACUGCAU .....(..((((...(((((.-...))))).....(((((--(..........(((.((((((....)))))))))((((-.....))))))))))..--....))-))..).. ( -35.90, z-score = -2.98, R) >droSec1.super_6 1710463 95 - 4358794 AUUGAGUGGUUAGAAUGGCCU-UUUGGUCGGCAGAACUAA--GACUGUAUACUCGUAGGUGGCACAGGUCACCACGGAGC------GCUCUUUAG---------UA-ACUGCAU ...............(((((.-...)))))((((.(((((--((.(((....((((.((((((....)))))))))).))------).)).))))---------).-.)))).. ( -28.30, z-score = -0.80, R) >droYak2.chr3R 17555824 103 - 28832112 CUUAAGGGAAAAGAAUGGCCUCUUUGCCCUUAAAAAC----------CAAAGGUAUAAGUGACACAGGUCACGCCGAAGCUAUAUAGUAUUAAAGAACUCCAAGUAACUUGCG- .(((((((.(((((......))))).)))))))....----------....(((....(((((....))))))))...(((....(((........)))...)))........- ( -21.00, z-score = -0.57, R) >droEre2.scaffold_4770 1868377 99 - 17746568 CUUGAGGGGAAAGAACGGCCUCUUUGGCCCUGGGAAC----------UAUAGCUAUAGGUGGCGCAGGUCACAAUGGAGC-AUAUAGCUAUGUAGGAU--CUGGUGCAUUGC-- ...((((((.........))))))..((.(..((..(----------((((((((((.(((((....)))))........-.)))))))..))))..)--)..).)).....-- ( -27.80, z-score = 0.45, R) >consensus CUUGAGUGGUUAGAAUGGCCU_UUUGGCCGGCAGAAC_AA___A___UAUACUCGUAGGUGGCACAGGUCACCACGGAGC_AUACAGCUCUUUAGUAU______UA_ACUGC__ ................((((.....))))............................((((((....))))))..(((((......)))))....................... (-15.82 = -16.10 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:10 2011