| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,596,385 – 1,596,475 |
| Length | 90 |
| Max. P | 0.972913 |

| Location | 1,596,385 – 1,596,475 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 65.44 |
| Shannon entropy | 0.63706 |
| G+C content | 0.47803 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -12.90 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
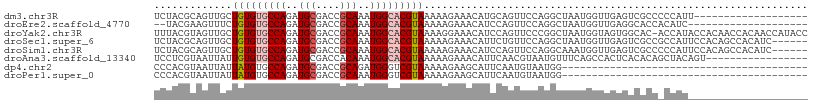
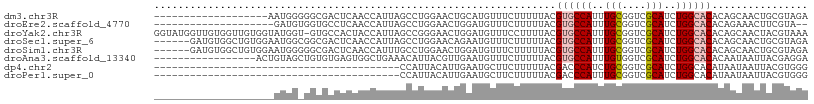
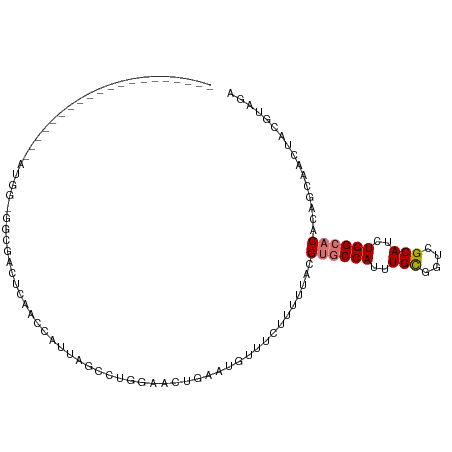
>dm3.chr3R 1596385 90 + 27905053 UCUACGCAGUUGCUGUGUGCCAGAUGCGACCGCAAAUGGCACGUAAAAAGAAACAUGCAGUUCCAGGCUAAUGGUUGAGUCGCCCCCAUU------------------- .....((.(..(((((((((((..(((....)))..))))))((........))..)))))..).((((........)))))).......------------------- ( -26.10, z-score = -0.89, R) >droEre2.scaffold_4770 1863058 87 + 17746568 --UACGAAGUUUCUGUGUGCCAGAUGCGACCGCAAAUGGCACGUAAAAAGAAACAUCCAGUUCCAGGCUAAUGGUUGAGGCACCACAUC-------------------- --...((.((((((.(((((((..(((....)))..))))))).....)))))).))..((..(((.(....).)))..))........-------------------- ( -27.00, z-score = -2.38, R) >droYak2.chr3R 17550703 108 + 28832112 UUUACGUAGUUGCUGUGUGCCAGAUGCGACCGCAAAUGGCACGUAAAAGGAAACAUCCAGUUCCCGGCUAAUGGUAGUGGCAC-ACCAUACCACAACCACAACCAUACC .....((((..((((.((((((..(((....)))..))))))......(....)...))))..).((....((((.((((...-......)))).))))...)).))). ( -32.20, z-score = -1.65, R) >droSec1.super_6 1705460 103 + 4358794 UCUACGCAGUUGCUGUGUGCCAGAUGCGACCGCAAAUGGCACGUAAAAAGAAACAUUCUGUUCCAGGCUAAUGGUUGAGUCGCCGCCAUUCCACAGCCACAUC------ ........((.(((((((((((..(((....)))..)))))).......................((..((((((.(.....).))))))))))))).))...------ ( -30.30, z-score = -1.20, R) >droSim1.chr3R 1627593 103 + 27517382 UCUACGCAGUUGCUGUGUGCCAGAUGCGACCGCAAAUGGCACGUAAAAAGAAACAUCCAGUUCCAGGCAAAUGGUUGAGUCGCCCCCAUUCCACAGCCACAUC------ ........((.(((((((((((..(((....)))..)))))).......((..((.(((............))).))..))...........))))).))...------ ( -28.20, z-score = -1.40, R) >droAna3.scaffold_13340 5192744 92 - 23697760 UCCUCGUAAUUAUUGUGUGCCAGAUGCGACCACAAAUGGCACGUAAAAAGAAACAUUCAACGUAAUGUUUCAGCCACUCACACAGCUACAGU----------------- .....(((....((((((((((..((......))..))))))))))...((((((((......))))))))...............)))...----------------- ( -20.30, z-score = -1.54, R) >dp4.chr2 17426309 68 + 30794189 CCCACGUAAUUAUUAUGUGCCAGAUGCGACCGCAGAUGGGUCGUAAAAAGAAGCAUUCAAUGUAAUGG----------------------------------------- ..((((((.....))))))(((..(((((((.......))))))).......((((...))))..)))----------------------------------------- ( -15.50, z-score = -1.07, R) >droPer1.super_0 11272987 68 + 11822988 CCCACGUAAUUAUUAUGUGCCAGAUGCGACCGCAAAUGGGUCGUAAAAAGAAGCAUUCAAUGUAAUGG----------------------------------------- ..((((((.....))))))(((..(((((((.......))))))).......((((...))))..)))----------------------------------------- ( -15.50, z-score = -1.20, R) >consensus UCUACGUAGUUGCUGUGUGCCAGAUGCGACCGCAAAUGGCACGUAAAAAGAAACAUUCAGUUCCAGGCUAAUGGUUGAGUCACC_CCAU____________________ .............(((((((((..(((....)))..)))))))))................................................................ (-12.90 = -13.34 + 0.44)
| Location | 1,596,385 – 1,596,475 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 65.44 |
| Shannon entropy | 0.63706 |
| G+C content | 0.47803 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.36 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1596385 90 - 27905053 -------------------AAUGGGGGCGACUCAACCAUUAGCCUGGAACUGCAUGUUUCUUUUUACGUGCCAUUUGCGGUCGCAUCUGGCACACAGCAACUGCGUAGA -------------------.....((((.............))))(((((.....)))))..(((((((((((..(((....)))..))))))...((....))))))) ( -27.92, z-score = -1.06, R) >droEre2.scaffold_4770 1863058 87 - 17746568 --------------------GAUGUGGUGCCUCAACCAUUAGCCUGGAACUGGAUGUUUCUUUUUACGUGCCAUUUGCGGUCGCAUCUGGCACACAGAAACUUCGUA-- --------------------...(((((......))))).............((.((((((......((((((..(((....)))..))))))..)))))).))...-- ( -24.90, z-score = -1.16, R) >droYak2.chr3R 17550703 108 - 28832112 GGUAUGGUUGUGGUUGUGGUAUGGU-GUGCCACUACCAUUAGCCGGGAACUGGAUGUUUCCUUUUACGUGCCAUUUGCGGUCGCAUCUGGCACACAGCAACUACGUAAA .(..((((((((((((((((((((.-...))).))))).)))))((((((.....))))))......((((((..(((....)))..))))))...)))))))..)... ( -44.40, z-score = -3.12, R) >droSec1.super_6 1705460 103 - 4358794 ------GAUGUGGCUGUGGAAUGGCGGCGACUCAACCAUUAGCCUGGAACAGAAUGUUUCUUUUUACGUGCCAUUUGCGGUCGCAUCUGGCACACAGCAACUGCGUAGA ------((((((((((..((.(((((((.............))).(((((.....))))).........))))))..))))))))))....((.(((...))).))... ( -35.12, z-score = -1.33, R) >droSim1.chr3R 1627593 103 - 27517382 ------GAUGUGGCUGUGGAAUGGGGGCGACUCAACCAUUUGCCUGGAACUGGAUGUUUCUUUUUACGUGCCAUUUGCGGUCGCAUCUGGCACACAGCAACUGCGUAGA ------.....(((.((((....(((....)))..))))..))).(((((.....)))))..(((((((((((..(((....)))..))))))...((....))))))) ( -35.80, z-score = -1.48, R) >droAna3.scaffold_13340 5192744 92 + 23697760 -----------------ACUGUAGCUGUGUGAGUGGCUGAAACAUUACGUUGAAUGUUUCUUUUUACGUGCCAUUUGUGGUCGCAUCUGGCACACAAUAAUUACGAGGA -----------------.......((.(((((((((..((((((((......))))))))...))))((((((..(((....)))..)))))).......))))).)). ( -26.50, z-score = -1.37, R) >dp4.chr2 17426309 68 - 30794189 -----------------------------------------CCAUUACAUUGAAUGCUUCUUUUUACGACCCAUCUGCGGUCGCAUCUGGCACAUAAUAAUUACGUGGG -----------------------------------------((((...((((..((((........((((((....).))))).....))))..))))......)))). ( -12.62, z-score = -0.65, R) >droPer1.super_0 11272987 68 - 11822988 -----------------------------------------CCAUUACAUUGAAUGCUUCUUUUUACGACCCAUUUGCGGUCGCAUCUGGCACAUAAUAAUUACGUGGG -----------------------------------------((((...((((..((((........((((((....).))))).....))))..))))......)))). ( -12.62, z-score = -0.66, R) >consensus ____________________AUGG_GGCGACUCAACCAUUAGCCUGGAACUGAAUGUUUCUUUUUACGUGCCAUUUGCGGUCGCAUCUGGCACACAGCAACUACGUAGA ...................................................................((((((..(((....)))..))))))................ (-12.97 = -13.36 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:08 2011