| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,549,316 – 1,549,420 |
| Length | 104 |
| Max. P | 0.961163 |

| Location | 1,549,316 – 1,549,420 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.34 |
| Shannon entropy | 0.49885 |
| G+C content | 0.47405 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -16.12 |
| Energy contribution | -15.18 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
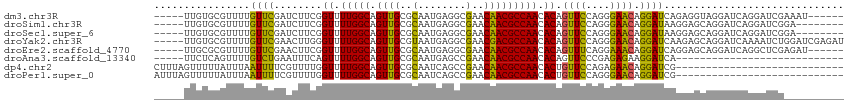
>dm3.chr3R 1549316 104 + 27905053 -----UUGUGCGUUUUGUUCGAUCUUCGGUUUUGGCAGUUGCGCAAUGAGGCGAACAACGCCAACACAGUUCCAGGGAACAGGAUCAGAGGUAGGAUCAGGAUCGAAAU------ -----............(((((((((..((.(((((.(((((((......)))..))))))))).)).((((....))))..((((........)))))))))))))..------ ( -32.50, z-score = -1.96, R) >droSim1.chr3R 1586234 102 + 27517382 -----UUGUGCGUUUUGUUCGAUCUUCGGUUUUGGCAGUUGCGCAAUGAGGCGAACAACGCCAACACAGUUCCAGGGAACAGGAUAAGGAGCAGGAUCAGGAUCGGA-------- -----((.((.(((((((((.(((((..((.(((((.(((((((......)))..))))))))).)).((((....)))))))))...))))))))))).)).....-------- ( -31.60, z-score = -1.51, R) >droSec1.super_6 1664976 102 + 4358794 -----UUGUGCGUUUUGUUCGAUCUUCGGUUUUGGCAGUUGCGCAAUGAGGCGAACAACGCCAACACAGUUCCAGGGAACAGGAUAAGGAGCAGGAUCAGGAUCGGA-------- -----((.((.(((((((((.(((((..((.(((((.(((((((......)))..))))))))).)).((((....)))))))))...))))))))))).)).....-------- ( -31.60, z-score = -1.51, R) >droYak2.chr3R 17513384 110 + 28832112 -----UUGUGCGUUUUGUUCGAACUUGGGUUUUGGCAGUUGCGUAAUGAGGCGAACGACGCCAACACAGUUCCAGGGAACAGGAUCAAGAGCAGGAUCAAAAUCUGGAUCGAGAU -----...(((((.((((..((((....))))..))))..)))))....((((.....))))......(.((((((......((((........))))....)))))).)..... ( -29.50, z-score = -0.35, R) >droEre2.scaffold_4770 1823683 104 + 17746568 -----UUGCGCGUUUUGUUCGAACUUCGGUUUUGGCAGUUGCGCAAUGAGGCGAACAACGCCAACACAGUUUCAGGAAACAGGAUCAGGAGCAGGAUCAGGCUCGAGAU------ -----(((((((..((((..((((....))))..)))).)))))))...((((.....))))......(((((.(....)........((((........)))))))))------ ( -35.30, z-score = -2.45, R) >droAna3.scaffold_13340 5153137 82 - 23697760 -----UUCUCAGUUUUGUCUGAAUUUCAGUUUUGGCAGUUGCGCAAUGAGCCGAACAACGCCAACACAGUUCCCGAGAGAAGGAUCA---------------------------- -----(((((.(..(((((((.....)))..(((((.((((.((.....))....))))))))).))))..)..)))))........---------------------------- ( -17.40, z-score = 0.25, R) >dp4.chr2 17388095 87 + 30794189 CUUUAGUUUUUAUUUAAUUUUCGUUUUGGUUUUGGCAGUUGCGCAAUCAGCCGAACAACGCCAACACUGUUCCAGAGAACAGGAUCG---------------------------- ...........................(((((((((.((((.((.....))....)))))))))..((((((....)))))))))).---------------------------- ( -17.90, z-score = -0.87, R) >droPer1.super_0 11234381 87 + 11822988 AUUUAGUUUUUAUUUAAUUUUCGUUUUGGUUUUGGCAGUUGCGCAAUCAGCCGAACAACGCCAACACUGUUCCAGGGAACAGGAUCG---------------------------- ...........................(((((((((.((((.((.....))....)))))))))..((((((....)))))))))).---------------------------- ( -19.80, z-score = -1.33, R) >consensus _____UUGUGCGUUUUGUUCGAACUUCGGUUUUGGCAGUUGCGCAAUGAGGCGAACAACGCCAACACAGUUCCAGGGAACAGGAUCAGGAGCAGGAUCAGGAUCG__________ ................((((........((.(((((.((((..(........)..))))))))).)).((((....)))).)))).............................. (-16.12 = -15.18 + -0.95)

| Location | 1,549,316 – 1,549,420 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.34 |
| Shannon entropy | 0.49885 |
| G+C content | 0.47405 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.65 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
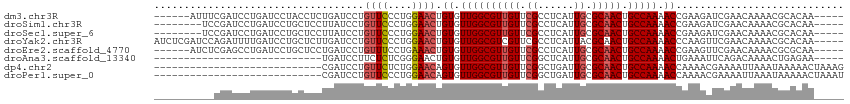
>dm3.chr3R 1549316 104 - 27905053 ------AUUUCGAUCCUGAUCCUACCUCUGAUCCUGUUCCCUGGAACUGUGUUGGCGUUGUUCGCCUCAUUGCGCAACUGCCAAAACCGAAGAUCGAACAAAACGCACAA----- ------..(((((((..((((........))))..((((....)))).((.(((((((((..(((......))))))).))))).))....)))))))............----- ( -26.90, z-score = -2.76, R) >droSim1.chr3R 1586234 102 - 27517382 --------UCCGAUCCUGAUCCUGCUCCUUAUCCUGUUCCCUGGAACUGUGUUGGCGUUGUUCGCCUCAUUGCGCAACUGCCAAAACCGAAGAUCGAACAAAACGCACAA----- --------..(((((....................((((....)))).((.(((((((((..(((......))))))).))))).))....)))))..............----- ( -23.30, z-score = -1.53, R) >droSec1.super_6 1664976 102 - 4358794 --------UCCGAUCCUGAUCCUGCUCCUUAUCCUGUUCCCUGGAACUGUGUUGGCGUUGUUCGCCUCAUUGCGCAACUGCCAAAACCGAAGAUCGAACAAAACGCACAA----- --------..(((((....................((((....)))).((.(((((((((..(((......))))))).))))).))....)))))..............----- ( -23.30, z-score = -1.53, R) >droYak2.chr3R 17513384 110 - 28832112 AUCUCGAUCCAGAUUUUGAUCCUGCUCUUGAUCCUGUUCCCUGGAACUGUGUUGGCGUCGUUCGCCUCAUUACGCAACUGCCAAAACCCAAGUUCGAACAAAACGCACAA----- .....((((.(((............))).))))..((((....))))((((((((((.....))))......((.((((...........)))))).....))))))...----- ( -21.90, z-score = -0.52, R) >droEre2.scaffold_4770 1823683 104 - 17746568 ------AUCUCGAGCCUGAUCCUGCUCCUGAUCCUGUUUCCUGAAACUGUGUUGGCGUUGUUCGCCUCAUUGCGCAACUGCCAAAACCGAAGUUCGAACAAAACGCGCAA----- ------...((((((..((((........))))..((((....)))).((.(((((((((..(((......))))))).))))).))....)))))).............----- ( -22.60, z-score = -0.48, R) >droAna3.scaffold_13340 5153137 82 + 23697760 ----------------------------UGAUCCUUCUCUCGGGAACUGUGUUGGCGUUGUUCGGCUCAUUGCGCAACUGCCAAAACUGAAAUUCAGACAAAACUGAGAA----- ----------------------------.........((((((....(((.((((((((((...((.....))))))).)))))..(((.....))))))...)))))).----- ( -23.70, z-score = -2.05, R) >dp4.chr2 17388095 87 - 30794189 ----------------------------CGAUCCUGUUCUCUGGAACAGUGUUGGCGUUGUUCGGCUGAUUGCGCAACUGCCAAAACCAAAACGAAAAUUAAAUAAAAACUAAAG ----------------------------((...((((((....))))))..((((((((((...((.....))))))).)))))........))..................... ( -19.90, z-score = -1.93, R) >droPer1.super_0 11234381 87 - 11822988 ----------------------------CGAUCCUGUUCCCUGGAACAGUGUUGGCGUUGUUCGGCUGAUUGCGCAACUGCCAAAACCAAAACGAAAAUUAAAUAAAAACUAAAU ----------------------------((...((((((....))))))..((((((((((...((.....))))))).)))))........))..................... ( -21.00, z-score = -2.32, R) >consensus __________CGAUCCUGAUCCUGCUCCUGAUCCUGUUCCCUGGAACUGUGUUGGCGUUGUUCGCCUCAUUGCGCAACUGCCAAAACCGAAGAUCGAACAAAACGCACAA_____ ...................................((((....)))).((.((((((((((.((......)).))))).))))).))............................ (-14.37 = -14.65 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:05 2011