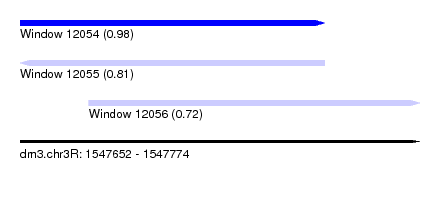
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,547,652 – 1,547,774 |
| Length | 122 |
| Max. P | 0.979973 |

| Location | 1,547,652 – 1,547,745 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Shannon entropy | 0.42081 |
| G+C content | 0.43299 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -11.41 |
| Energy contribution | -11.73 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.979973 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
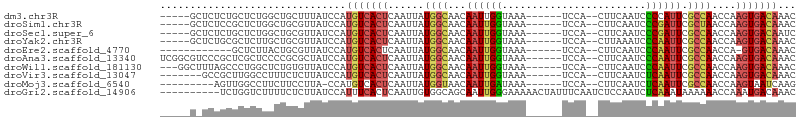
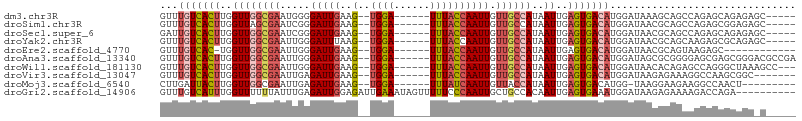
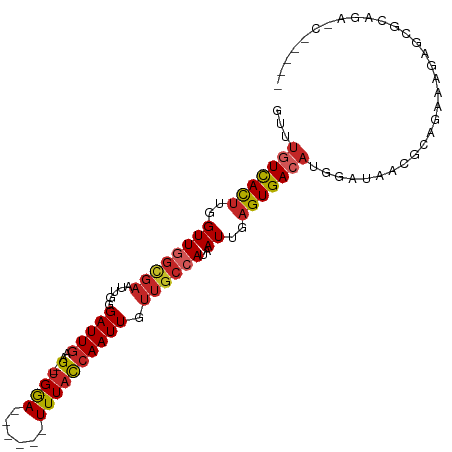
>dm3.chr3R 1547652 93 + 27905053 -----GCUCUCUGCUCUGGCUGCUUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCCAUUCGCCAACCAAGUGACAAAC -----((..((......))..))........(((((((......((....)).((((..((------(...--...........)))..))))...)))))))... ( -16.14, z-score = -1.28, R) >droSim1.chr3R 1584571 93 + 27517382 -----GCUCUCCGCUCUGGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCGAUUCGCUAACCAAGUGACAAAC -----......(((.......))).......(((((((......((....)).((((..((------((..--..........))))..))))...)))))))... ( -18.20, z-score = -1.60, R) >droSec1.super_6 1663321 93 + 4358794 -----GCUCUCUGCUCUGGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCGAUUCGCCAACCAAGUGACAAUC -----((..((......))..))........(((((((......((....)).((((..((------((..--..........))))..))))...)))))))... ( -20.00, z-score = -2.06, R) >droYak2.chr3R 17511746 93 + 28832112 -----GCUCUGCGCUCUUGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUAAAUCCCAAUUCGCCAACCAAGUGACAAAC -----.....((((.......))))......(((((((......((((...((((((....------....--........)))))).))))....)))))))... ( -20.69, z-score = -2.82, R) >droEre2.scaffold_4770 1822065 85 + 17746568 ------------GCUCUUACUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCAAUUCGCCAACCA-GUGACAAAC ------------((.......))........(((((((......((((...((((((....------....--........)))))).))))...)-))))))... ( -17.29, z-score = -3.08, R) >droAna3.scaffold_13340 5151487 98 - 23697760 UCGGCGUCCCGCUCGCUCCCCGCGCUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCAAUUCGCCAACCAAGUGACAAAC ..((((...))))(((.....))).......(((((((......((((...((((((....------....--........)))))).))))....)))))))... ( -20.69, z-score = -2.41, R) >droWil1.scaffold_181130 7779464 95 - 16660200 ---GGCUUUAGCCCUGGCUCUGUGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCAAUUCGCCAACCAAGUGACAAAC ---(((....))).((((.....))))....(((((((......((((...((((((....------....--........)))))).))))....)))))))... ( -20.69, z-score = -1.63, R) >droVir3.scaffold_13047 12653319 91 - 19223366 -------GCCGCUUGGCCUUUCUCUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCUCAAUUCGCCAACCAAGUGACAAAC -------(((....)))..............(((((((......((....)).((((..((------(...--...........)))..))))...)))))))... ( -17.44, z-score = -2.66, R) >droMoj3.scaffold_6540 32639798 88 - 34148556 ---------AGUUGGCCUUCUUCCUUA-CCAUGUCACUCAAUUAUGGUAACAAUUGAUAAA------UCCA--CUUCAAUCUCAAUUCGCCAACCAAGUAAUCAAG ---------.((((((........(((-(((((.........))))))))..(((((....------....--..)))))........))))))............ ( -17.10, z-score = -4.27, R) >droGri2.scaffold_14906 2310204 96 - 14172833 ----------UCUGGUCUUUUCUCUUAUCCAUUUCACUCAAUUGUGGCAGCAAUUGGGAAAAACUAUUUCAAUCUCCAAUCUCAAAUAAAAAACCAAAUGACAAAC ----------..((((.((((................((((((((....))))))))((((.....)))).................)))).)))).......... ( -11.50, z-score = -0.62, R) >consensus _____G_UCUGCGCUCUUGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA______UCCA__CUUCAAUCCCAAUUCGCCAACCAAGUGACAAAC ...............................(((((((......((((...((((((........................)))))).))))....)))))))... (-11.41 = -11.73 + 0.32)
| Location | 1,547,652 – 1,547,745 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Shannon entropy | 0.42081 |
| G+C content | 0.43299 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -15.55 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806629 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
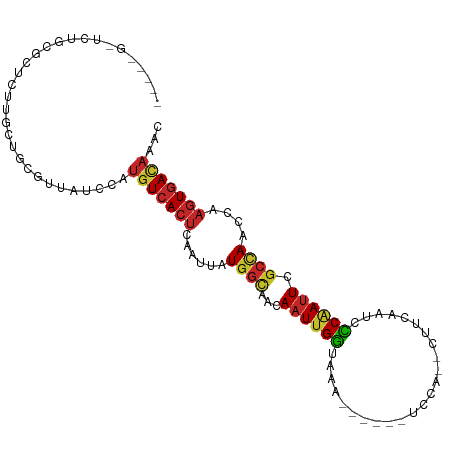
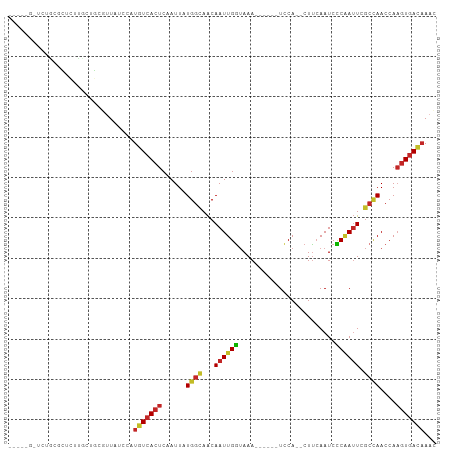
>dm3.chr3R 1547652 93 - 27905053 GUUUGUCACUUGGUUGGCGAAUGGGGAUUGAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAAAGCAGCCAGAGCAGAGAGC----- ((((((((((..((((((((.((((((((....--..))------))).)))....))))))..))..)))))).(((.........))))))).......----- ( -26.20, z-score = -1.98, R) >droSim1.chr3R 1584571 93 - 27517382 GUUUGUCACUUGGUUAGCGAAUCGGGAUUGAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGCCAGAGCGGAGAGC----- ...(((((((..((((((((....((..((((.--....------)))))).....))))..))))..))))))).......(((.......)))......----- ( -23.20, z-score = -0.73, R) >droSec1.super_6 1663321 93 - 4358794 GAUUGUCACUUGGUUGGCGAAUCGGGAUUGAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGCCAGAGCAGAGAGC----- ...(((((((..((((((((....((..((((.--....------)))))).....))))))..))..)))))))........((.......)).......----- ( -23.70, z-score = -0.94, R) >droYak2.chr3R 17511746 93 - 28832112 GUUUGUCACUUGGUUGGCGAAUUGGGAUUUAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGCAAGAGCGCAGAGC----- ...(((((((..((((((((((((((((((...--.)))------))..)))))..))))))..))..)))))))........((.((....)))).....----- ( -28.10, z-score = -2.42, R) >droEre2.scaffold_4770 1822065 85 - 17746568 GUUUGUCAC-UGGUUGGCGAAUUGGGAUUGAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGUAAGAGC------------ ...((((((-(.(.(((((((((((........--....------....)))))..))))))....).)))))))...................------------ ( -20.99, z-score = -1.20, R) >droAna3.scaffold_13340 5151487 98 + 23697760 GUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAGCGCGGGGAGCGAGCGGGACGCCGA ...(((((((..(((((((((((((........--....------....)))))..))))))..))..))))))).......(((.....)))..(((....))). ( -30.89, z-score = -1.75, R) >droWil1.scaffold_181130 7779464 95 + 16660200 GUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACACAGAGCCAGGGCUAAAGCC--- ...(((((((..(((((((((((((........--....------....)))))..))))))..))..)))))))(((...........))).(((....)))--- ( -27.89, z-score = -1.83, R) >droVir3.scaffold_13047 12653319 91 + 19223366 GUUUGUCACUUGGUUGGCGAAUUGAGAUUGAAG--UGGA------UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAAGAGAAAGGCCAAGCGGC------- ...(((((((..((((((((.....(((((...--....------.....))))).))))))..))..)))))))..............(((....)))------- ( -25.00, z-score = -2.25, R) >droMoj3.scaffold_6540 32639798 88 + 34148556 CUUGAUUACUUGGUUGGCGAAUUGAGAUUGAAG--UGGA------UUUAUCAAUUGUUACCAUAAUUGAGUGACAUGG-UAAGGAAGAAGGCCAACU--------- ...........(((((((..(((........))--)...------(((((((..((((((((....)).)))))))))-)))).......)))))))--------- ( -19.60, z-score = -1.53, R) >droGri2.scaffold_14906 2310204 96 + 14172833 GUUUGUCAUUUGGUUUUUUAUUUGAGAUUGGAGAUUGAAAUAGUUUUUCCCAAUUGCUGCCACAAUUGAGUGAAAUGGAUAAGAGAAAAGACCAGA---------- ........(((((((((((.(((..((((((.((..((.....))..)))))))).....(((......))).........))).)))))))))))---------- ( -18.10, z-score = -0.37, R) >consensus GUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAG__UGGA______UUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGAAAGAGCGCAGA_C_____ ...(((((((..((((((((.....(((((....................))))).))))))..))..)))))))............................... (-15.55 = -16.39 + 0.84)
| Location | 1,547,673 – 1,547,774 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Shannon entropy | 0.42244 |
| G+C content | 0.44610 |
| Mean single sequence MFE | -18.61 |
| Consensus MFE | -10.86 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
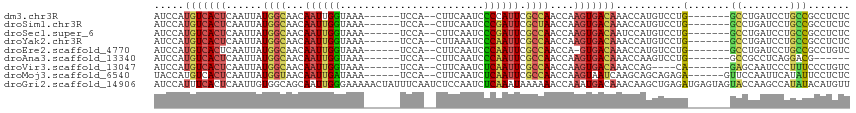
>dm3.chr3R 1547673 101 + 27905053 AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCCAUUCGCCAACCAAGUGACAAACCAUGUCCUG-------GCCUGAUCCUGCCGCCUCUC .....(((((((......((....)).((((..((------(...--...........)))..))))...)))))))...........(-------((........)))....... ( -17.94, z-score = -1.15, R) >droSim1.chr3R 1584592 101 + 27517382 AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCGAUUCGCUAACCAAGUGACAAACCAUGUCCUG-------GCCUGAUCCUGCCGCCUCUC .....(((((((......((....)).((((..((------((..--..........))))..))))...)))))))...........(-------((........)))....... ( -19.00, z-score = -1.37, R) >droSec1.super_6 1663342 101 + 4358794 AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCGAUUCGCCAACCAAGUGACAAUCCAUGUCCUG-------GCCUGAUCCUGCCGCCUCUC .....(((((((......((....)).((((..((------((..--..........))))..))))...)))))))...........(-------((........)))....... ( -21.30, z-score = -2.00, R) >droYak2.chr3R 17511767 101 + 28832112 AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUAAAUCCCAAUUCGCCAACCAAGUGACAAACCAUGUCCUG-------GCCUGAUCCUGCCGCCUCUC .....(((((((......((((...((((((....------....--........)))))).))))....)))))))...........(-------((........)))....... ( -20.09, z-score = -2.14, R) >droEre2.scaffold_4770 1822079 100 + 17746568 AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCAAUUCGCCAACCA-GUGACAAACCAUGUCCUG-------GCCUGAUCCUGCCGCCUGUC .....(((((((......((((...((((((....------....--........)))))).))))...)-))))))...........(-------((........)))....... ( -19.69, z-score = -1.21, R) >droAna3.scaffold_13340 5151513 95 - 23697760 AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCAAGUCCUG-------GCCGCCUCAGGACG------ .....(((((((......((((...((((((....------....--........)))))).))))....)))))))......((((((-------(.....))))))).------ ( -26.59, z-score = -4.42, R) >droVir3.scaffold_13047 12653338 97 - 19223366 AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA------UCCA--CUUCAAUCUCAAUUCGCCAACCAAGUGACAAACCAG----CA-------GAGCAAUCCCUUUCCCUGUC .....(((((((......((....)).((((..((------(...--...........)))..))))...))))))).....(----((-------(.(.((.....)).))))). ( -15.84, z-score = -1.64, R) >droMoj3.scaffold_6540 32639814 102 - 34148556 UACCAUGUCACUCAAUUAUGGUAACAAUUGAUAAA------UCCA--CUUCAAUCUCAAUUCGCCAACCAAGUAAUCAAGCAGCAGAGA------GUUCCAAUUCAUAUUCCUCUC .....((..((((.....((((...((((((....------....--........)))))).)))).....((......))......))------))..))............... ( -10.39, z-score = -0.46, R) >droGri2.scaffold_14906 2310220 116 - 14172833 AUCCAUUUCACUCAAUUGUGGCAGCAAUUGGGAAAAACUAUUUCAAUCUCCAAUCUCAAAUAAAAAACCAAAUGACAAACAAGCUGAGAUGAGUAGUACCAAGCCAUAUACAUGUU ..........(((((((((....)))))))))....(((((((.........((((((..........................)))))))))))))................... ( -16.67, z-score = 0.14, R) >consensus AUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAA______UCCA__CUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCAUGUCCUG_______GCCUGAUCCUGCCGCCUCUC .....(((((((......((((...((((((........................)))))).))))....)))))))....................................... (-10.86 = -11.25 + 0.39)
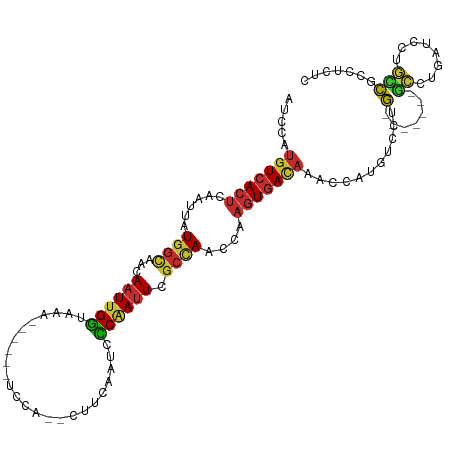
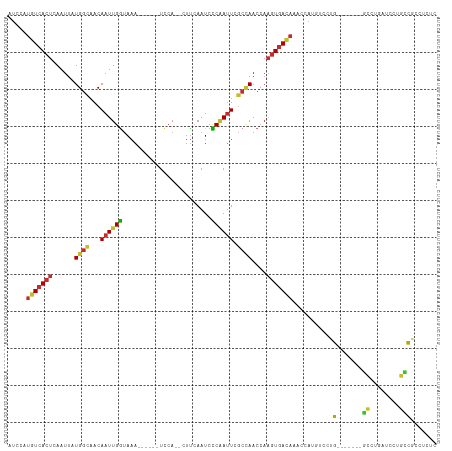
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:03 2011