| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,393,616 – 6,393,733 |
| Length | 117 |
| Max. P | 0.567031 |

| Location | 6,393,616 – 6,393,733 |
|---|---|
| Length | 117 |
| Sequences | 11 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 62.20 |
| Shannon entropy | 0.79480 |
| G+C content | 0.39229 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -9.03 |
| Energy contribution | -8.89 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
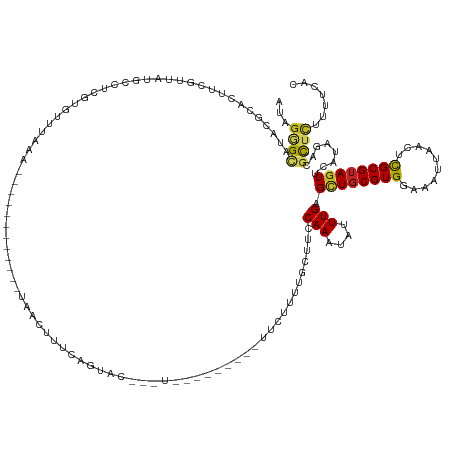
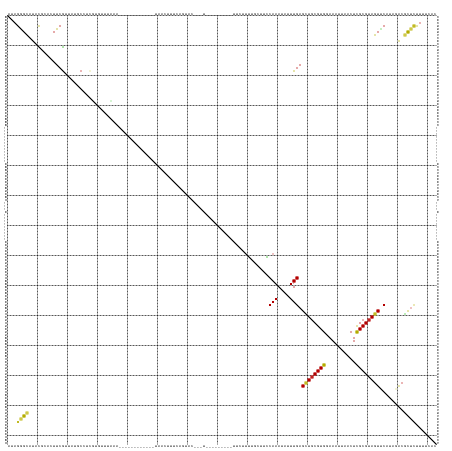
>dm3.chr2L 6393616 117 - 23011544 AUAGGGCAUACGCAAUUCGUUAUGCUUUGUGUUUCA-------------AAACUUUCAGUAC-------------UUCUUGUGCUUCCAAAUAUUUGAGCUGCGUGGAAAUUAACUCGCGUAGUUCAUAAGCGCUUUUUUCAC ..((((((((((.....)).))))))))((((((..-------------........(((((-------------.....)))))..........((((((((((((........)))))))))))).))))))......... ( -31.80, z-score = -2.84, R) >droSim1.chr2L 6197781 118 - 22036055 UUAGGGCAUCCACACUUCGUUAUGCUUUGUGUUUUAA------------AAACUUUCAGUAC-------------UUCUUGUGCUGCCAAAUAUUUGAGCUCCGUGGAAAUUAACUCGCGUAGUUCAUAAGCGCUUUUUUCAC ..(((((((..((.....)).)))))))((((((...------------.......((((((-------------.....)))))).........((((((.(((((........))))).)))))).))))))......... ( -27.30, z-score = -1.68, R) >droSec1.super_5 4461376 118 - 5866729 UUAGGGCAUCCACACUUCGUUAUGCUUCGUGUUUUAA------------AAACUUUCAGUAC-------------UUCUUGUGCUGCCAAAUAUUUGAGCUGCGUGGAAAUUAACUCGCGUAGUCCAUAAGCGCUUUUUUCAC ...((((((..((.....)).)))))).((((((...------------.......((((((-------------.....)))))).........((..((((((((........))))))))..)).))))))......... ( -27.60, z-score = -1.26, R) >droYak2.chr2L 15813427 118 + 22324452 AUAGGGCAUACGCACUUCGAUAUACUUCGUGUUUCAA------------AAACUUUCAGUAC-------------UUCUUGCGCUGCCAAAUAUUUGAGCUGCGUGAAAAUUAACUCGCGUAGUUCAUAAGCGUUUUUUUCAC ....((((..((((....((..((((....(((....------------.)))....)))).-------------.)).)))).)))).......((((((((((((........))))))))))))................ ( -30.30, z-score = -2.35, R) >droEre2.scaffold_4929 15319471 118 - 26641161 AAAGGGCAUACGCACUUCGUUGUCCCUCGUGUUUCAC------------AAACUUUCAGUGC-------------UUCUUGUGCUGCCAAAUAUUUGAGCUGCGUGGAAAUUAACUCGCGUAGUUCAUAAGCUCUUUUUUCAC ((((((((((((.((......))....))))).....------------.......(((..(-------------.....)..))).........((((((((((((........))))))))))))...)))))))...... ( -31.30, z-score = -2.14, R) >droAna3.scaffold_12916 8327862 120 + 16180835 ---GGGCGUUCAGAAUUCG-UAUGCCAUC--UUAACGCUUUGCAGUCUUU-------UUUUGCCCUGGG-------CGUUU---AUACAAAUAUUUGAGGUGCGUGGAAAUUAACUCGCGUAGUUCUUAGACUUACUUUUUGC ---.(((((...(....).-.)))))...--..(((((((.((((.....-------..))))...)))-------)))).---............(((.(((((((........))))))).)))................. ( -27.20, z-score = -0.05, R) >dp4.chr4_group5 480178 136 + 2436548 AUAGAGCAUACCCACUUCG-UGCUCCUCUAUUUAAUUUCAUUCUGUUUUUUGUGUUUUUUUUUCCUCCG---UUUUCGUCU---GUGCAAAUAUUUGAGCUGCGUGUAAAUUAACUCGCGUAGUGCUGCGACGCUCCUUUUAA ...((((((.........)-)))))..........................(((((.............---...(((..(---((....)))..)))((((((((..........)))))))).....)))))......... ( -21.20, z-score = -0.23, R) >droPer1.super_5 451466 134 + 6813705 AUAGAGCAUACCCACUUCG-UGCUCCUCUAUUUAAUUUCAUUCUGUUUUUUGUGU--UUUUUUCCUCCG---UUUUCGUCU---GUGCAAAUAUUUGAGCUGCGUGUAAAUUAACUCGCGUAGUGCUGCGACGCUCCUUUUAA ...((((((.........)-)))))..........................((((--(...........---...(((..(---((....)))..)))((((((((..........)))))))).....)))))......... ( -21.20, z-score = -0.19, R) >droWil1.scaffold_180708 5513187 143 + 12563649 UUAGGGCAUUUGAAAUUCACCAUACCUAAAAUUUGAAUCCCUUUUGUUGUAAUUUUUAAUCCCUUUUAGCUGUUAUUUUUUUUCUUGCAAAUAUUUGAGCUGCGUGGAAAUUAACUUGCGUAGUUCGUCGAGGCUCUUUUUAA ..(((((.(((((.................(((((.............(((((..((((......))))..)))))...........)))))....((((((((..(........)..)))))))).))))))))))...... ( -26.85, z-score = -0.78, R) >droVir3.scaffold_12963 17504411 140 + 20206255 UAAGAGUAUUCGAGCUUCGAUAUGCGAAUAGCUUUUUGCGUUUCUUAAUUCUUUUUAACUGCUGCUGCU---GCUUUUCUUGGCGUACAAAUAUUUGAGCUGCGUGUAAAUUAACUCGCGUAGUUCUUCGACGCUGCUUUAAC ..(((((.....(((.((((((((((((.(((.....(((.(..((((......))))..).)))....---))).)))...))))).........((((((((((..........)))))))))).)))).))))))))... ( -33.60, z-score = -0.25, R) >droGri2.scaffold_15252 14878282 134 + 17193109 CCAGAGUAUUUCAGCUUCGGUAUGCGAAUGGUUAAUUUCGUAUCUUAAUUCUUUUUAACUGCUGCUA---------UUCUUGGCGUGCAAAUAUUUGAGUUGCGUGUAAAUUAACUCGCGUAGUUCUUCGACGCCCCUUUAAC .............((.(((((((((((...((((((((((((.(((((.......((..((((((((---------....))))).)))..)).))))).))))...))))))))))))))).....)))).))......... ( -28.50, z-score = -0.44, R) >consensus AUAGGGCAUACGCACUUCGUUAUGCCUCGUGUUUAAA____________UAACUUUCAGUAC___U_________UUCUUUUGCUUCCAAAUAUUUGAGCUGCGUGGAAAUUAACUCGCGUAGUUCAUAGACGCUCUUUUCAC ...((((................................................................................(((....))).((((((((..........))))))))........))))....... ( -9.03 = -8.89 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:11 2011