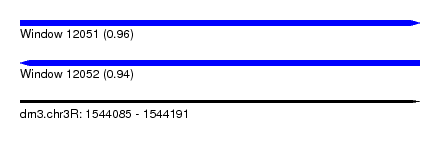
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,544,085 – 1,544,191 |
| Length | 106 |
| Max. P | 0.959783 |

| Location | 1,544,085 – 1,544,191 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.01 |
| Shannon entropy | 0.48509 |
| G+C content | 0.45678 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -19.59 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
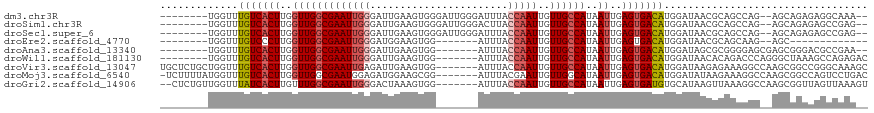
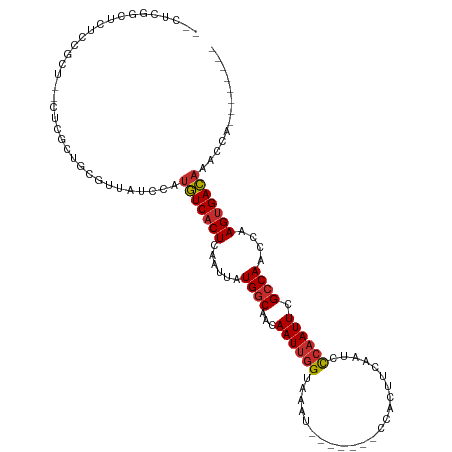
>dm3.chr3R 1544085 106 + 27905053 --------UGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGGGAUUGGGAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGCCAG--AGCAGAGAGGCAAA-- --------((((((((((((..(((((((((((........)))..((((((......)))))).))))))..))..)))))))..........))))).--.((......))...-- ( -29.80, z-score = -1.66, R) >droSim1.chr3R 1580958 106 + 27517382 --------UGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGGGAUUGGGACUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGCCAG--AGCAGAGAGCCGAG-- --------.(((((((((((..(((((((((((((.....((((.........)))).)))))..))))))..))..)))))).........((......--.))...)))))...-- ( -31.60, z-score = -1.87, R) >droSec1.super_6 1659710 106 + 4358794 --------UGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGGGAUUGGGAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGCCAG--AGCAGAGAGCCGAG-- --------.(((((((((((..(((((((((((........)))..((((((......)))))).))))))..))..)))))).........((......--.))...)))))...-- ( -31.50, z-score = -2.07, R) >droEre2.scaffold_4770 1818438 88 + 17746568 --------UGGUUUGUCCCUUGGUUGGCGAAUUGGGAUGGAAGUGG-------AUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGCAAG--AGC------------- --------.....((((.((..(((((((((((((....(((....-------.))).)))))..))))))..))..)).))))................--...------------- ( -20.10, z-score = 0.17, R) >droAna3.scaffold_13340 5147829 101 - 23697760 --------UGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGG-------AUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAGCGCGGGGAGCGAGCGGGACGCCGAA-- --------.(((.(((((((..(((((((((((((...........-------.....)))))..))))))..))..))))))).......(((.(....)..)))....)))...-- ( -30.99, z-score = -1.65, R) >droWil1.scaffold_181130 7775683 103 - 16660200 --------UGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGG-------AUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACACAGACCCAGGGCUAAAGCCAGAGAC --------.(((((((((((..(((((((((((((...........-------.....)))))..))))))..))..)))))).((.....)).)))))...(((....)))...... ( -31.49, z-score = -2.30, R) >droVir3.scaffold_13047 12649250 111 - 19223366 UGCUCUGCUGGUUUGUCACUUGGUUGGCGAAUUGAGAUUGAAGUGG-------AUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAAGAGAAAGGCCAAGCGGCCGGGCAAAGC .(((.((((....(((((((..((((((((.....(((((......-------......))))).))))))..))..))))))).............((((....)))).)))).))) ( -37.60, z-score = -2.79, R) >droMoj3.scaffold_6540 32636373 110 - 34148556 -UCUUUUAUGGUUUGUCACUUGGUUGGCGAAUGGAGAUGGAAGCGG-------AUUUACGAAUUGUUGGCAUAAUUGAGUGACAUGGAUAUAAGAAAGGCCAAGCGGCCAGUCCUGAC -..(((((((...(((((((..((((((.((..(.....(((....-------.))).....)..)).)).))))..)))))))....)))))))..((((....))))......... ( -32.40, z-score = -2.14, R) >droGri2.scaffold_14906 2307346 109 - 14172833 --CUCUGUUGGUUUAUCACUUGUUUGGCGAAUUGGGACUAAAGUGG-------AUUUACCAAUUGUUGCCAUAAUUGAGUGAUGUGCAUAAGUUAAAGGCCAAGCGGUUAGUUAAAGU --..((((((((((((((((((..(((((((((((...........-------.....)))))..))))))....))))))))..((....))...))))).)))))........... ( -25.39, z-score = -0.48, R) >consensus ________UGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGG_______AUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGACAG__AGCGGAGAGCCGAA__ .............(((((((..(((((((((((((.......................)))))..))))))..))..))))))).................................. (-19.59 = -20.08 + 0.48)
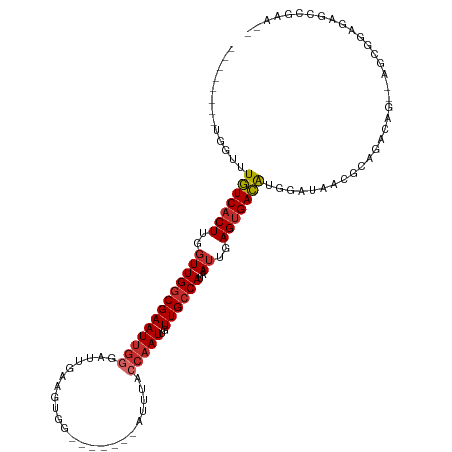
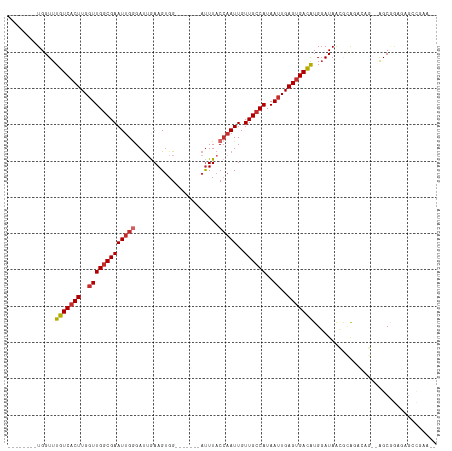
| Location | 1,544,085 – 1,544,191 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Shannon entropy | 0.48509 |
| G+C content | 0.45678 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1544085 106 - 27905053 --UUUGCCUCUCUGCU--CUGGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUCCCAAUCCCACUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCA-------- --...(((........--..)))...........(((((((......((((...((((((.......................)))))).))))....))))))).....-------- ( -19.50, z-score = -1.72, R) >droSim1.chr3R 1580958 106 - 27517382 --CUCGGCUCUCUGCU--CUGGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAGUCCCAAUCCCACUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCA-------- --(.((((.(......--..))))).).......(((((((......((((...((((((.((((.........)))).....)))))).))))....))))))).....-------- ( -20.10, z-score = -1.09, R) >droSec1.super_6 1659710 106 - 4358794 --CUCGGCUCUCUGCU--CUGGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUCCCAAUCCCACUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCA-------- --(.((((.(......--..))))).).......(((((((......((((...((((((.......................)))))).))))....))))))).....-------- ( -19.50, z-score = -1.32, R) >droEre2.scaffold_4770 1818438 88 - 17746568 -------------GCU--CUUGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAU-------CCACUUCCAUCCCAAUUCGCCAACCAAGGGACAAACCA-------- -------------.((--((((..(((.......(((((........)))))..((((((.....-------...........)))))))))....))))))........-------- ( -14.39, z-score = -0.25, R) >droAna3.scaffold_13340 5147829 101 + 23697760 --UUCGGCGUCCCGCUCGCUCCCCGCGCUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAU-------CCACUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCA-------- --...((((...))))(((.....))).......(((((((......((((...((((((.....-------...........)))))).))))....))))))).....-------- ( -20.69, z-score = -2.30, R) >droWil1.scaffold_181130 7775683 103 + 16660200 GUCUCUGGCUUUAGCCCUGGGUCUGUGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAU-------CCACUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCA-------- (.(((.(((....)))..))).)...........(((((((......((((...((((((.....-------...........)))))).))))....))))))).....-------- ( -23.49, z-score = -1.57, R) >droVir3.scaffold_13047 12649250 111 + 19223366 GCUUUGCCCGGCCGCUUGGCCUUUCUCUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAU-------CCACUUCAAUCUCAAUUCGCCAACCAAGUGACAAACCAGCAGAGCA (((((((..((((....)))).............(((((((......((....)).((((..(((-------..............)))..))))...))))))).....))))))). ( -32.44, z-score = -4.28, R) >droMoj3.scaffold_6540 32636373 110 + 34148556 GUCAGGACUGGCCGCUUGGCCUUUCUUAUAUCCAUGUCACUCAAUUAUGCCAACAAUUCGUAAAU-------CCGCUUCCAUCUCCAUUCGCCAACCAAGUGACAAACCAUAAAAGA- ...((((..((((....))))..)))).......(((((((...(((((.........)))))..-------..((..............))......)))))))............- ( -17.94, z-score = -0.65, R) >droGri2.scaffold_14906 2307346 109 + 14172833 ACUUUAACUAACCGCUUGGCCUUUAACUUAUGCACAUCACUCAAUUAUGGCAACAAUUGGUAAAU-------CCACUUUAGUCCCAAUUCGCCAAACAAGUGAUAAACCAACAGAG-- ............((((((((...........))..............((((...((((((((((.-------....))))...)))))).))))..))))))..............-- ( -15.30, z-score = -0.55, R) >consensus __CUCGGCUCUCCGCU__CUCGCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAU_______CCACUUCAAUCCCAAUUCGCCAACCAAGUGACAAACCA________ ..................................(((((((......((((...((((((.......................)))))).))))....)))))))............. (-13.16 = -13.34 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:52:00 2011