| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,534,180 – 1,534,282 |
| Length | 102 |
| Max. P | 0.516768 |

| Location | 1,534,180 – 1,534,282 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.23 |
| Shannon entropy | 0.76002 |
| G+C content | 0.51278 |
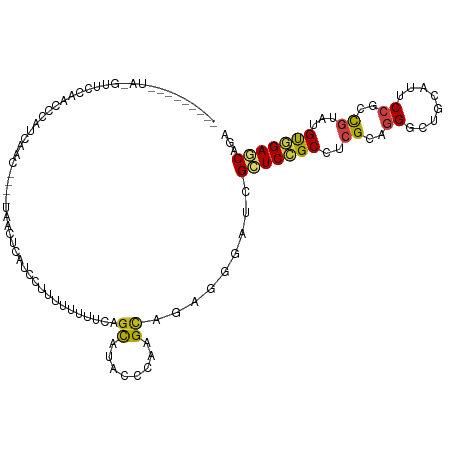
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.26 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
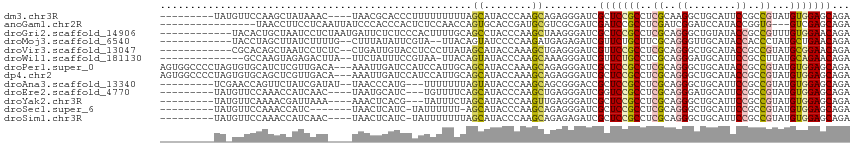
>dm3.chr3R 1534180 102 + 27905053 ---------UAUGUUCCAAGCUAUAAAC----UAACGCACCCUUUUUUUUUAGCAUACCCAAGCAGAGGGAUCGCUCCGCCUCGCAAGGCUGCAUUCCGCCGUAUGUGGAGCAGA ---------..((((((..((.......----....))..............((((((....((.(((((.......).))))))..(((........))))))))))))))).. ( -26.10, z-score = -0.42, R) >anoGam1.chr2R 14625799 96 - 62725911 ----------------UAACCUUCCUCAAUUAUCCCACCCACUCUCCAACCAGUGCACCGAUGCGUCGCGAUCGAUCCGCCUCGAUCGGAUCCAUACCGGUG---GUCGAGCAGA ----------------....((..(((............((((........))))((((((((.(((.(((((((......)))))))))).)))..)))))---...))).)). ( -25.90, z-score = -1.52, R) >droGri2.scaffold_14906 11852313 103 + 14172833 ------------UACACUGCUAAUCCUCUAAUGAUUCUCUCCCACUUUUGCAGCCUACCCAAGCUAAGGGAUCGCUCCGCCUCGCAGGGCUGUAUACCGCCGUUUGUGGAACAGA ------------....(((.............................((((((((.(((.......)))...((........)).))))))))..((((.....))))..))). ( -23.80, z-score = 0.17, R) >droMoj3.scaffold_6540 32626061 99 - 34148556 ------------UACCUAGCUUAUCUUUUG--CUUUAUAUUCGUA--UUACAGUAUCCCCAAGAUGAGAGAUCGUUCUGCUUCGCAGGGUUGCAUACCACCCUAUGCUGAACAGA ------------.......((((((((.((--((..(((....))--)...)))).....)))))))).....((((.((.....(((((........)))))..)).))))... ( -19.70, z-score = -0.47, R) >droVir3.scaffold_13047 6557660 101 + 19223366 ------------CGCACAGCUAAUCCUCUC--CUGAUUGUACCUCCCUUAUAGCAUACCAAAGCUGAGGGAUCGUUCCGCCUCGCAGGGCUGCAUACCGCCGUAUGCGGAACAGA ------------......((.((((.....--..))))))...((((((..(((........)))))))))..(((((((((....)))).((((((....)))))))))))... ( -31.00, z-score = -1.29, R) >droWil1.scaffold_181130 7765484 98 - 16660200 --------------GCCAAGUAGAGACUUA--UUCUAUUUCCGUAA-UUACAGUAUACCCAAGCAAAGGGAUCGUUCUGCCUCGCAGGGAUGCAUUCCGCCUUAUGCAGAACAGA --------------...((((((((.....--))))))))......-..........(((.......)))...(((((((...((.((((....)))))).....)))))))... ( -21.80, z-score = -0.41, R) >droPer1.super_0 11218555 112 + 11822988 AGUGGCCCCUAGUGUGCAUCUCGUUGACA---AAAUUGAUCCAUCCAUUGCAGCAUACCAAAGCAGAGGGAUCGCUCCGCCUCGCAGGGCUGCAUACCGCCGUAUGUGGAGCAGA ...(..((((..(((.......((((...---.(((.((....)).))).))))........))).))))..)(((((((((....)))).((((((....)))))))))))... ( -32.76, z-score = 0.62, R) >dp4.chr2 17372462 112 + 30794189 AGUGGCCCCUAGUGUGCAGCUCGUUGACA---AAAUUGAUCCAUCCAUUGCAGCAUACCAAAGCAGAGGGAUCGCUCCGCCUCGCAGGGCUGCAUACCGCCGUAUGUGGAGCAGA ...(..((((.((((((.((.........---.................)).))))))........))))..)(((((((((....)))).((((((....)))))))))))... ( -33.97, z-score = 0.57, R) >droAna3.scaffold_13340 5138110 101 - 23697760 ---------UCGAACCAGUUCUAUCGAUAU--UAACCCAUG---UUUUUUUAGUAUACCCAAGCAGCGGGACCGCUCCGCCUCGCAGGGCUGCAUUCCGCCGUAUGUGGAGCAGA ---------........(((((((..((((--(((......---.....)))))))......((.((((((..((...((((....)))).)).)))))).))..)))))))... ( -24.70, z-score = 0.35, R) >droEre2.scaffold_4770 1808642 99 + 17746568 ---------UAUGUUCCAAACCAUCAAC----UAAUGCAUC---UGUUUUCAGCAUACCCAAGCUGAGGGAUCGGUCCGCCUCGCAGGGAUGCAUUCCGCCGUAUGUGGAGCAGA ---------..(((((((..........----.((((((((---(...((((((........))))))(((....))).........)))))))))..........))))))).. ( -30.10, z-score = -1.02, R) >droYak2.chr3R 17498048 99 + 28832112 ---------UAUGUUCAAAACGAUUAAA----AAACUCACG---UAUUUCUAGCAUACCCAAGUUGAGGGAUCGCUCCGCCUCGCAGGGCUGCAUUCCGCCGUAUGUGGAGCAGA ---------............((((...----.((((...(---(((.......))))...))))....))))(((((((((....)))).((((........)))))))))... ( -23.10, z-score = 0.32, R) >droSec1.super_6 1649873 97 + 4358794 ---------UAUGUUCCAAACCAUC-------UAACUCAUC-UAUUUUUU-AGCAUACCCAAGCAGAGGGAUCGCUCCGCCUCGCAGGGCUGCAUUCCGCCGUAUGUGGAGCAGA ---------..(((((((.......-------........(-((.....)-))(((((....((.(((((.......).))))))..(((........))))))))))))))).. ( -23.90, z-score = -0.09, R) >droSim1.chr3R 1568715 101 + 27517382 ---------UAUGUUCCAAACCAUCAAC----UAACUCAUC-UAUUUUUUUAGCAUACCCAAGCAGAGAGAUCGCUCCGCCUCGCAGGGCUGCAUUCCGCCGUAUGUGGAGCAGA ---------..(((((((..........----........(-((......)))(((((....((.(((......))).)).......(((........))))))))))))))).. ( -21.70, z-score = -0.14, R) >consensus _________UA_GUUCCAACCCAUCAAC____UAACUCAUCCUUUUUUUUCAGCAUACCCAAGCAGAGGGAUCGCUCCGCCUCGCAGGGCUGCAUUCCGCCGUAUGUGGAGCAGA ....................................................((........)).........(((((((..((..((........))..))...)))))))... (-12.46 = -12.34 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:59 2011