
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,488,626 – 1,488,716 |
| Length | 90 |
| Max. P | 0.828872 |

| Location | 1,488,626 – 1,488,716 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 58.20 |
| Shannon entropy | 0.76901 |
| G+C content | 0.37919 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -7.01 |
| Energy contribution | -6.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.828872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
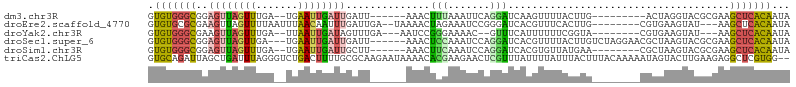
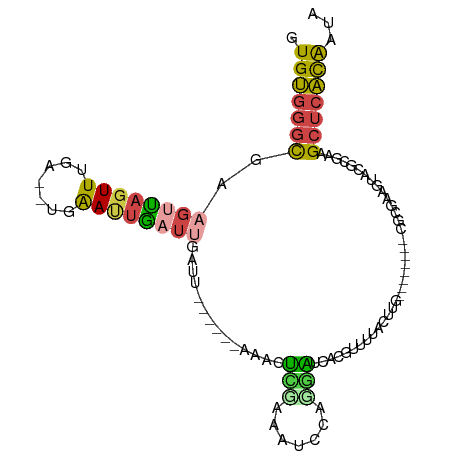
>dm3.chr3R 1488626 90 - 27905053 GUGUGGGCGGAGUUAGUUUGA--UGAAUUGAUUGAUU------AAACUUUAAAUUCAGGAUCAAGUUUUACUUG---------ACUAGGUACGCGAAGCUCACAAUA .(((((((.((((((((((((--(.((....)).)))------)))))...))))).((.(((((.....))))---------)))...........)))))))... ( -24.30, z-score = -2.77, R) >droEre2.scaffold_4770 1762428 94 - 17746568 GUGUGCGCGAAGUUAGUUUUAAUUUAACAAUUUGAUUGA--UAAAACUAGAAAUCCGGGAUCACGUUUCACUUG--------CGUGAAGUAU---AAGCUCACAAUA .((((.((((..(((((((((..((((....))))....--)))))))))...)).....((((((.......)--------))))).....---..)).))))... ( -19.90, z-score = -1.01, R) >droYak2.chr3R 17455508 89 - 28832112 GUGUGGGCGAAGUUAGUUUGA--UUAAUUGAUAGUUUGA---AAUCCGGGAAAAC--GUUUCAUUUUUUCGGUA--------CGUGAAGUAU---AAGCUCACAAUA .(((((((..(((((((...)--)))))).(((.((((.---...(((..((((.--......))))..)))..--------..)))).)))---..)))))))... ( -21.90, z-score = -2.19, R) >droSec1.super_6 1603462 98 - 4358794 GUGUGGGCGGAGUUAGUUGA---UGAAUUGAUUGAUU------AAACUCCAAAUCCAGGAUCACGUUUUACUUGUCUAGGAACGCUAAGUACGCGAAGCUCACAAUA .((((((((((((((((..(---(......))..)))------.))))))...(((.(((.((.(.....).))))).))).(((.......)))..)))))))... ( -30.80, z-score = -3.04, R) >droSim1.chr3R 1523447 91 - 27517382 GUGUGGGCGGAGUUAGUUUGA--UGAAUUGAUUGCUU------AAACUUCAAAUCCAGGAUCACGUGUUAUGAA--------CGCUAAGUACGCGAAGCUCACAAUA .((((((((((...(((((((--............))------))))).....))).......(((((......--------........)))))..)))))))... ( -22.04, z-score = -0.68, R) >triCas2.ChLG5 1685134 105 - 18847211 GUGCAGAUUAGCUGAUUUAGGGUCUGACUUUUGCGCAAGAAUAAAACACGAAGAACUCGUUUAUUUUAUUUACUUUACAAAAAUAGUACUUGAAGAGGCUCGUGG-- (((((((....(.(((.....))).)...)))))))..........(((((....(((.((((..((((((.........))))))....)))))))..))))).-- ( -20.00, z-score = -0.08, R) >consensus GUGUGGGCGAAGUUAGUUUGA__UGAAUUGAUUGAUU______AAACUCGAAAUCCAGGAUCACGUUUUACUUG________CGCGAAGUACGCGAAGCUCACAAUA .(((((((..((((((((.......))))))))..............(((.......))).....................................)))))))... ( -7.01 = -6.82 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:53 2011