
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,424,419 – 1,424,514 |
| Length | 95 |
| Max. P | 0.998942 |

| Location | 1,424,419 – 1,424,514 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 65.47 |
| Shannon entropy | 0.72632 |
| G+C content | 0.32961 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -11.72 |
| Energy contribution | -10.99 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.998942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
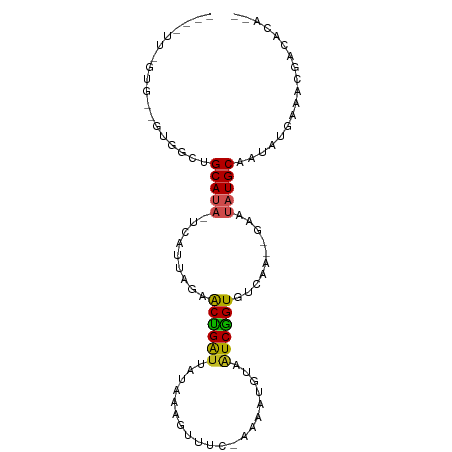
>dm3.chr3R 1424419 95 - 27905053 GAAAUUCGUGAAGUGGCUGCAUAUC-AUUAGAGCUGACUAUAAAGUUUCAAAAAUGUAAUCGGUGUCAAGAAU--AUGCAAUAACAAUCAACACAUUU-- .......(((...((..(((((((.-.((...(((((.((((............)))).)))))...))..))--)))))....)).....)))....-- ( -15.50, z-score = -0.15, R) >droSim1.chr3R 1460767 94 - 27517382 GUAAUUCGUGAAGUGGCAGCAUAUC-AUUAGAGCUGAUUAUAAAGUUUU-AAAAUGUAAUCGGUGUCAAGAAU--AUGCAAUAGCAAUAGACACAUUU-- .......(((.....((.((((((.-.((...((((((((((.......-....))))))))))...))..))--))))....))......)))....-- ( -18.70, z-score = -1.29, R) >droSec1.super_6 1539480 94 - 4358794 GUAAUUCGUGAAGUGGCUGCAUAUC-AUUAGAGCUGAUUAUAAAGUUUU-AAAAUGUAAUCAGUGUCAAGAAU--AUGCAAUAACAAUCGACACAUUU-- .....(((....((...(((((((.-.((...((((((((((.......-....))))))))))...))..))--)))))...))...))).......-- ( -20.00, z-score = -1.71, R) >droYak2.chr3R 17389105 77 - 28832112 -----------------UGCAUAUU-AUUAGAACCGAUUAUAAAGUUUC-AAAAUGUUAUCGGUGUCCAAAAU--AUGCAAUAUAAAUCGGCACAUUU-- -----------------((((((((-......((((((.(((.......-....))).))))))......)))--)))))..................-- ( -15.00, z-score = -2.14, R) >droEre2.scaffold_4770 1694827 82 - 17746568 ------------GUGGUUGCAUAUC-AUUAGAGCUGAUUAUAAAGUUUU-AAAAUGUAAUCGGUGUCAAGAAU--AUGCAAUAUCAAUUGACACAUUU-- ------------((((((((((((.-.((...((((((((((.......-....))))))))))...))..))--))))))).((....)))))....-- ( -21.40, z-score = -3.29, R) >droAna3.scaffold_13088 68277 78 + 569066 ----------------CUGCAUAUU-AUUAGAACUGGUUGUAAAGUUCA-AAAAGACAAUCAGUGUCGAAAAU--AUGCAAUAAGAAACGACUCAUUU-- ----------------.((((((((-.((...(((((((((........-.....)))))))))...)).)))--)))))..................-- ( -18.02, z-score = -3.20, R) >dp4.chr2 7424691 94 + 30794189 UUUGUGUGUGUGUUGUGUGCAUAUA-AUUAGAACUGAUUACAAGCUCA--AAAAUGUAGUCGGUGUC-AGAUU--AUGCCACAUGGAACGCCACAUAUUU ..((((((.((((((((((..((((-(((.((((((((((((......--....)))))))))).))-.))))--))).)))))..)))))))))))... ( -31.20, z-score = -3.36, R) >droPer1.super_3 1234757 94 + 7375914 UUUGUGUGUGUGUUGUGUGCAUAUA-AUUAGAACUGAUUACAAGCUCA--AAAAUGUAGUCGGUGUC-AGAUU--AUGCCACAUGGAACGCCACAUAUUU ..((((((.((((((((((..((((-(((.((((((((((((......--....)))))))))).))-.))))--))).)))))..)))))))))))... ( -31.20, z-score = -3.36, R) >droWil1.scaffold_181108 3226697 94 + 4707319 -----UUGUUUGUUUUUUGCAUAUUAAUUAGAACCGGUUAUACAAAUUCAUAUAUUUUAUCGGUGUCAAUAUAUUAUGCCAAAUGAAACGCCACGUUUU- -----..((((...(((.(((((.(((((.((((((((((((........))))....)))))).))))).)).))))).)))..))))..........- ( -12.70, z-score = 0.39, R) >droVir3.scaffold_13047 11661766 88 + 19223366 -----CAAUGCUGCGUGUGCAUAUU-AUUAGAACCGGUUAU--CGGUCAAUGCAGAUAAUCGGUGUCAUUAUA---UGCCAAAUGAAACGCCUCAUUUU- -----.((((..((((..((((((.-......(((((((((--(.((....)).))))))))))......)))---)))........))))..))))..- ( -24.82, z-score = -2.13, R) >droGri2.scaffold_14830 2853045 90 + 6267026 -----GAAAACCUUCGCCGCACAUU-AUUAGAACCGGUUAGUUGUACUUGUA-AUAUAAUCGGUGUCAAAAUGU--UGCAAAAUGAAACGCCACAUUUU- -----.............(((((((-......((((((((....((....))-...)))))))).....)))).--)))((((((........))))))- ( -16.10, z-score = -0.38, R) >consensus _____U_GUG__GUGGCUGCAUAUU_AUUAGAACUGAUUAUAAAGUUUU_AAAAUGUAAUCGGUGUCAAGAAU__AUGCAAUAUGAAACGACACAUUU__ ..................((((..........(((((((((..............)))))))))...........))))..................... (-11.72 = -10.99 + -0.73)



| Location | 1,424,422 – 1,424,514 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 64.07 |
| Shannon entropy | 0.76934 |
| G+C content | 0.34228 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -8.98 |
| Energy contribution | -8.46 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1424422 92 - 27905053 GAAAUUCGUGAAGUGGCUGCAUA-UCAUUAGAGCUGACUAUAAAGUUUCAAAAAUGUAAUCGGUGUCAA--GAAUAUGCAAUAACAAUCAACACA-- .......(((...((..((((((-(..((...(((((.((((............)))).)))))...))--..)))))))....)).....))).-- ( -15.50, z-score = -0.27, R) >droSim1.chr3R 1460770 91 - 27517382 GUAAUUCGUGAAGUGGCAGCAUA-UCAUUAGAGCUGAUUAUAAAGUUUU-AAAAUGUAAUCGGUGUCAA--GAAUAUGCAAUAGCAAUAGACACA-- .......(((.....((.(((((-(..((...((((((((((.......-....))))))))))...))--..))))))....))......))).-- ( -18.70, z-score = -1.40, R) >droSec1.super_6 1539483 91 - 4358794 GUAAUUCGUGAAGUGGCUGCAUA-UCAUUAGAGCUGAUUAUAAAGUUUU-AAAAUGUAAUCAGUGUCAA--GAAUAUGCAAUAACAAUCGACACA-- .....(((....((...((((((-(..((...((((((((((.......-....))))))))))...))--..)))))))...))...)))....-- ( -20.00, z-score = -1.79, R) >droYak2.chr3R 17389108 74 - 28832112 -----------------UGCAUA-UUAUUAGAACCGAUUAUAAAGUUUC-AAAAUGUUAUCGGUGUCCA--AAAUAUGCAAUAUAAAUCGGCACA-- -----------------((((((-((......((((((.(((.......-....))).)))))).....--.))))))))...............-- ( -15.00, z-score = -2.21, R) >droEre2.scaffold_4770 1694830 79 - 17746568 ------------GUGGUUGCAUA-UCAUUAGAGCUGAUUAUAAAGUUUU-AAAAUGUAAUCGGUGUCAA--GAAUAUGCAAUAUCAAUUGACACA-- ------------(((((((((((-(..((...((((((((((.......-....))))))))))...))--..))))))))).((....))))).-- ( -21.40, z-score = -3.40, R) >droAna3.scaffold_13088 68280 75 + 569066 ----------------CUGCAUA-UUAUUAGAACUGGUUGUAAAGUUCA-AAAAGACAAUCAGUGUCGA--AAAUAUGCAAUAAGAAACGACUCA-- ----------------.((((((-((.((...(((((((((........-.....)))))))))...))--.))))))))...............-- ( -18.02, z-score = -3.34, R) >dp4.chr2 7424694 91 + 30794189 UUUGUGUGUGUGUUGUGUGCAUA-UAAUUAGAACUGAU--UACAAGCUCAAAAAUGUAGUCGGUGUCAG---AUUAUGCCACAUGGAACGCCACAUA ..((((.((((.(..((((..((-(((((.((((((((--((((..........)))))))))).)).)---)))))).))))..).)))))))).. ( -31.10, z-score = -3.40, R) >droPer1.super_3 1234760 91 + 7375914 UUUGUGUGUGUGUUGUGUGCAUA-UAAUUAGAACUGAU--UACAAGCUCAAAAAUGUAGUCGGUGUCAG---AUUAUGCCACAUGGAACGCCACAUA ..((((.((((.(..((((..((-(((((.((((((((--((((..........)))))))))).)).)---)))))).))))..).)))))))).. ( -31.10, z-score = -3.40, R) >droWil1.scaffold_181108 3226701 90 + 4707319 -----UUGUUUGUUUUUUGCAUAUUAAUUAGAACCGGUUAUACAAAUUCAUAUAUUUUAUCGGUGUCAAUAUAUUAUGCCAAAUGAAACGCCACG-- -----..((((...(((.(((((.(((((.((((((((((((........))))....)))))).))))).)).))))).)))..))))......-- ( -12.70, z-score = 0.09, R) >droVir3.scaffold_13047 11661770 89 + 19223366 GACACCAAUGCUGCGUGUGCAUA-UUAUUAGAACCGGUUAUCGGUCAAU-GCAG-AUAAUCGGUGUCAU--UAUA-UGCCAAAUGAAACGCCUCA-- .((((((....)).))))(((((-(.......((((((((((.((....-)).)-))))))))).....--.)))-)))................-- ( -23.52, z-score = -1.24, R) >droGri2.scaffold_14830 2853049 91 + 6267026 AAUUGGAAAACCUUCGCCGCACA-UUAUUAGAACCGGUUAGUUGUACUU-GUAAUAUAAUCGGUGUCAA--AAUGUUGCAAAAUGAAACGCCACA-- ...(((......((((..(((((-((......((((((((....((...-.))...)))))))).....--)))).)))....))))...)))..-- ( -16.00, z-score = 0.25, R) >consensus ____UU_GUG__GUGGCUGCAUA_UCAUUAGAACUGAUUAUAAAGUUUC_AAAAUGUAAUCGGUGUCAA__GAAUAUGCAAUAUGAAACGACACA__ ..................(((((.........((((((....................))))))..........))))).................. ( -8.98 = -8.46 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:46 2011