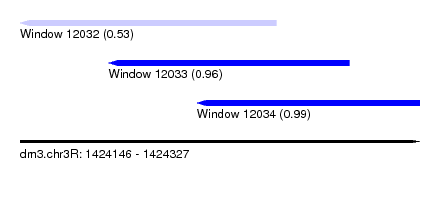
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,424,146 – 1,424,327 |
| Length | 181 |
| Max. P | 0.993951 |

| Location | 1,424,146 – 1,424,262 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Shannon entropy | 0.52893 |
| G+C content | 0.29416 |
| Mean single sequence MFE | -18.49 |
| Consensus MFE | -9.01 |
| Energy contribution | -8.51 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
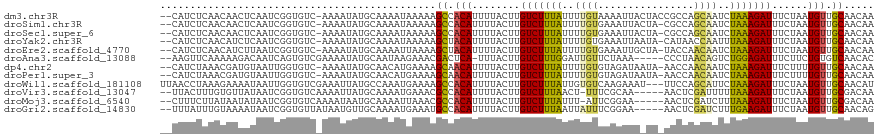
>dm3.chr3R 1424146 116 - 27905053 --CAUCUCAACAACUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCCACAUUUUACUUGUCUUUAUUUUGUAAAAUUACUACCGCCAGCAAUCUAAAGAUUUCUAAUGUUGCAACAA --.....(((((.......(((((..-......(((((((((((.....(((.......))).))))))))))).......)))))..((.((((....)))).))..)))))...... ( -16.96, z-score = -1.76, R) >droSim1.chr3R 1460492 115 - 27517382 --CAUCUCAACAACUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCCACAUUUUACUUGUCUUUAUUUUGUGAAAUUACUA-CGCCAGCAAUCUAAAGAUUUCUAAUGUUGCAACAA --.....(((((........(((((.-.......((((((((((.....(((.......))).)))))))))).........)-))))((.((((....)))).))..)))))...... ( -17.83, z-score = -1.37, R) >droSec1.super_6 1539209 115 - 4358794 --CAUCUCAACAACUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCCACAUUUUACUUGUCUUUAUUUUGUGAAAUUACUA-CGCCAGCAAUCUAAAGAUUUCUAAUGUUGCAACAA --.....(((((........(((((.-.......((((((((((.....(((.......))).)))))))))).........)-))))((.((((....)))).))..)))))...... ( -17.83, z-score = -1.37, R) >droYak2.chr3R 17388842 115 - 28832112 --CAUCUCAACAUCUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCUACAUUUUACUUGUCUUUAUUUUGUGAAAUUAAUA-CAUAACCAAUUUAAAGAUUUCUAAUGUUGCAACAA --.......(((((......))))).-...................((.(((((......(((((((..(((...........-......)))..)))))))....))))).))..... ( -15.43, z-score = -1.18, R) >droEre2.scaffold_4770 1694564 115 - 17746568 --CAUCUCAACAUCUUAAUCGGUGUC-AAAAUAUGCAAAAUUAAAAGCUACAUUUUACUUGUCUUUAUUUUGUGAAAUUGCUA-UACCAACAAUCUAAAGAUUUCUAAUGUUGCAACAA --.......(((((......))))).-...................((.(((((......(((((((..((((..........-.....))))..)))))))....))))).))..... ( -17.36, z-score = -1.50, R) >droAna3.scaffold_13088 68023 111 + 569066 --AAGUUCAAAAAGACAAUCAGUGUCGAAAAUAUGCAAUAAGAAACGACUCA-UUUACUUGUCUUUGGAUUGUUCUAAA-----CCCUAACAGUCUGGAGAUUUCUUCUGUGUCAACAC --..(((......((((.....))))....................(((.((-.......(((((..(((((((.....-----....)))))))..)))))......)).)))))).. ( -22.52, z-score = -0.84, R) >dp4.chr2 7424419 115 + 30794189 --CAUCUAAACGAUGUAAUUGGUGUC-AAAAUAUGCAACAUGAAAAGCAACAUUUUACUUGUCUUUAUUUUGUGUAGAUAAUA-AACCAACAAUCUAAAGAUUUCUUUUGUUGCAACAA --((((.....))))...((.((((.-..........)))).))..((((((........(((((((..((((..........-.....))))..)))))))......))))))..... ( -19.20, z-score = -0.58, R) >droPer1.super_3 1234486 115 + 7375914 --CAUCUAAACGAUGUAAUUGGUGUC-AAAAUAUGCAACAUGAAAAGCAACAUUUUACUUGUCUUUAUUUUGUGUAGAUAAUA-AACCAACAAUCUAAAGAUUUCUUUUGUUGCAACAA --((((.....))))...((.((((.-..........)))).))..((((((........(((((((..((((..........-.....))))..)))))))......))))))..... ( -19.20, z-score = -0.58, R) >droWil1.scaffold_181108 3226417 116 + 4707319 UUAACCUAAAGAAAAUAAUUGGUGUCGAAAUUAUGCCAAAUGAAAAGCCACAUUUUACUUGUCUUUAUUGUGUCAAGAAAU---UUCCAGCAUUCUAAAGAUUUCUAAUGUUGCAACAU ..................(((((((.......))))))).......((.(((((......(((((((..((((...((...---.))..))))..)))))))....))))).))..... ( -19.10, z-score = -0.96, R) >droVir3.scaffold_13047 11661514 111 + 19223366 --UUACUUUGUGUUAUAAUCGGUGUCAAAAUUAUGCAAAAUGAAACGCCACAUUUUACUUGUCUUUAACU-UUUCGCAA-----AACUCGAUUUUUAAAGAUUUCUAAUGUUGCGACAA --....................((((....((((.....))))...((.(((((......((((((((..-..(((...-----....)))...))))))))....))))).)))))). ( -17.40, z-score = -0.54, R) >droMoj3.scaffold_6540 30630503 111 + 34148556 --CUUUCUUAUAAUAUAAUCGGUGUCAAAAUAAUGCAAAAUUAAACGCCACAUUUUACUUGUCUUUAUUU-AUUCGGAA-----AACUCGAUCUUUAAAGAUUUCUAAUGUUGCGACAA --....................((((....((((.....))))...((.(((((......(((((((...-..((((..-----...))))....)))))))....))))).)))))). ( -18.70, z-score = -2.21, R) >droGri2.scaffold_14830 2852772 112 + 6267026 --UUUAUUUGUAAAAUAAUCGGUGUUAUAAUGUUGCAAAAUGAAAUGCCACAUUUUACUUGUCUUUAAUUAUUUCGGAA-----AACUCGAUCUUUGAAGAUUUCUAAUGUUGCAACAG --(((((((((((.(((((....)))))....)))).))))))).(((.(((((......((((((((.....((((..-----...))))...))))))))....))))).))).... ( -20.30, z-score = -1.40, R) >consensus __CAUCUCAACAACAUAAUCGGUGUC_AAAAUAUGCAAAAUGAAAAGCCACAUUUUACUUGUCUUUAUUUUGUGAAAAUA_UA_AACCAGCAAUCUAAAGAUUUCUAAUGUUGCAACAA ..............................................((.(((........(((((((..((((................))))..)))))))......))).))..... ( -9.01 = -8.51 + -0.50)
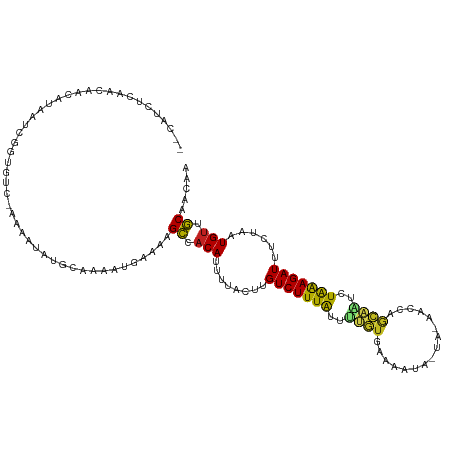
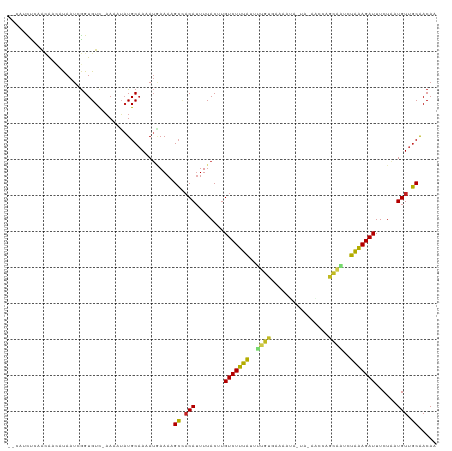
| Location | 1,424,186 – 1,424,295 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.55 |
| Shannon entropy | 0.55585 |
| G+C content | 0.29878 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -12.02 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
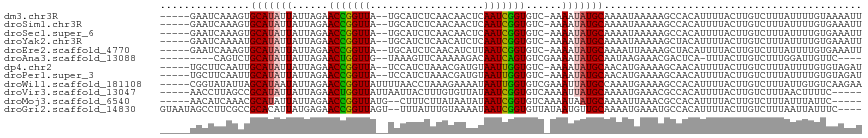
>dm3.chr3R 1424186 109 - 27905053 -----GAAUCAAAGUGCAUAUUAUUAGAACCGGUUA--UGCAUCUCAACAACUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCCACAUUUUACUUGUCUUUAUUUUGUAAAAUU -----.........((((((((......(((((((.--.(...........)..)))))))...-..))))))))..............(((((((.............))))))). ( -16.52, z-score = -1.01, R) >droSim1.chr3R 1460531 109 - 27517382 -----GAAUCAAAGUGCAUAUUAUUAGAACCGGUUA--UGCAUCUCAACAACUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCCACAUUUUACUUGUCUUUAUUUUGUGAAAUU -----.........((((((((......(((((((.--.(...........)..)))))))...-..))))))))..............(((((((.............))))))). ( -16.62, z-score = -0.52, R) >droSec1.super_6 1539248 109 - 4358794 -----GAAUCAAAGUGCAUAUUAUUAGAACCGGUUA--UGCAUCUCAACAACUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCCACAUUUUACUUGUCUUUAUUUUGUGAAAUU -----.........((((((((......(((((((.--.(...........)..)))))))...-..))))))))..............(((((((.............))))))). ( -16.62, z-score = -0.52, R) >droYak2.chr3R 17388881 109 - 28832112 -----GAAUCAAAAUGCAUAUUAUUAGAACCGGUUA--UGCAUCUCAACAUCUCAAUCGGUGUC-AAAAUAUGCAAAAUAAAAAGCUACAUUUUACUUGUCUUUAUUUUGUGAAAUU -----.........((((((((......((((((((--((........)))...)))))))...-..))))))))..............(((((((.............))))))). ( -17.92, z-score = -1.41, R) >droEre2.scaffold_4770 1694603 109 - 17746568 -----GAAUCAAAGUGCAUAUUAUUAGAACCGGUUA--UGCAUCUCAACAUCUUAAUCGGUGUC-AAAAUAUGCAAAAUUAAAAGCUACAUUUUACUUGUCUUUAUUUUGUGAAAUU -----.........((((((((......((((((((--...............))))))))...-..))))))))..............(((((((.............))))))). ( -18.78, z-score = -1.33, R) >droAna3.scaffold_13088 68062 101 + 569066 ---------CAGUCUGCAUAUUAUUAGAACUGGUUG--UAAAGUUCAAAAAGACAAUCAGUGUCGAAAAUAUGCAAUAAGAAACGACUCA-UUUACUUGUCUUUGGAUUGUUC---- ---------(((((((((((((.((...((((((((--(.............)))))))))...)).)))))))..........(((...-.......)))...))))))...---- ( -23.82, z-score = -2.25, R) >dp4.chr2 7424458 109 + 30794189 -----UGCUUCAAUUGCAUAUUAUUAGAACCGGUUA--UCCAUCUAAACGAUGUAAUUGGUGUC-AAAAUAUGCAACAUGAAAAGCAACAUUUUACUUGUCUUUAUUUUGUGUAGAU -----(((((((.(((((((((......((((((((--..((((.....))))))))))))...-..)))))))))..)))..))))....(((((...............))))). ( -24.36, z-score = -2.10, R) >droPer1.super_3 1234525 109 + 7375914 -----UGCUUCAAUUGCAUAUUAUUAGAACCGGUUA--UCCAUCUAAACGAUGUAAUUGGUGUC-AAAAUAUGCAACAUGAAAAGCAACAUUUUACUUGUCUUUAUUUUGUGUAGAU -----(((((((.(((((((((......((((((((--..((((.....))))))))))))...-..)))))))))..)))..))))....(((((...............))))). ( -24.36, z-score = -2.10, R) >droWil1.scaffold_181108 3226454 112 + 4707319 -----CGGUAUAUUAGCAUAAUAUUAGAACCGGUUAUUUUAACCUAAAGAAAAUAAUUGGUGUCGAAAUUAUGCCAAAUGAAAAGCCACAUUUUACUUGUCUUUAUUGUGUCAAGAA -----.(((.((((.(((((((.((...((((((((((((.........))))))))))))...)).)))))))..))))....)))........((((.(........).)))).. ( -23.20, z-score = -2.10, R) >droVir3.scaffold_13047 11661553 107 + 19223366 -----AACCUUAGCCGCAUAUUAUUAGAACUGGUUAUUAAUUACUUUGUGUUAUAAUCGGUGUCAAAAUUAUGCAAAAUGAAACGCCACAUUUUACUUGUCUUUAACUUUUC----- -----..........(((((........(((((((((.(((........))))))))))))........)))))((((((........))))))..................----- ( -16.09, z-score = -0.57, R) >droMoj3.scaffold_6540 30630542 105 + 34148556 -----AACAUCAAACGCAUAUUAUUAGAACCGGUUAUG--CUUUCUUAUAAUAUAAUCGGUGUCAAAAUAAUGCAAAAUUAAACGCCACAUUUUACUUGUCUUUAUUUAUUC----- -----..........((((.........((((((((((--.((......)))))))))))).........))))......................................----- ( -14.17, z-score = -1.12, R) >droGri2.scaffold_14830 2852811 111 + 6267026 GUAAUAGCCUUCGCCGCACAUUAUGAGAACCGGUUAGU--UUUAUUUGUAAAAUAAUCGGUGUUAUAAUGUUGCAAAAUGAAAUGCCACAUUUUACUUGUCUUUAAUUAUUUC---- .(((..((.......(((((((((((..((((((((.(--((((....))))))))))))).)))))))).)))((((((........))))))....))..)))........---- ( -23.80, z-score = -2.10, R) >consensus _____GAAUCAAAGUGCAUAUUAUUAGAACCGGUUA__UGCAUCUCAACAACAUAAUCGGUGUC_AAAAUAUGCAAAAUGAAAAGCCACAUUUUACUUGUCUUUAUUUUGUGAAAUU ...............(((((((......(((((((...................)))))))......)))))))........................................... (-12.02 = -12.00 + -0.01)
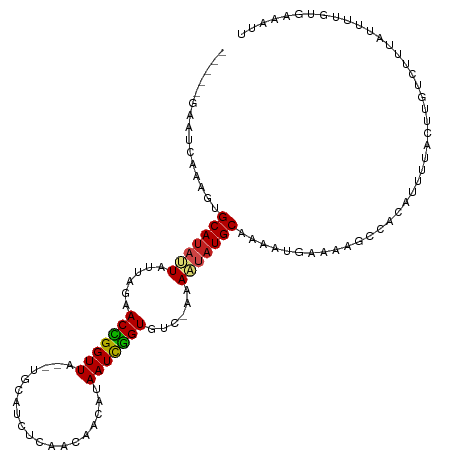
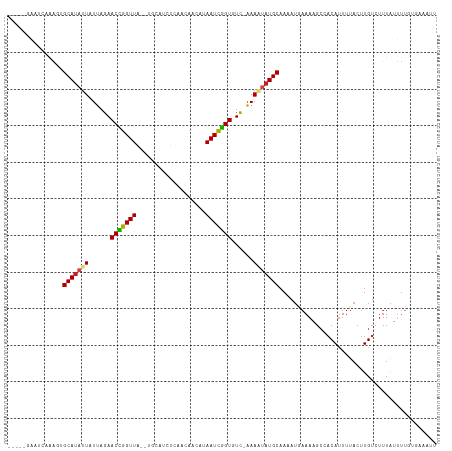
| Location | 1,424,226 – 1,424,327 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 61.11 |
| Shannon entropy | 0.86754 |
| G+C content | 0.34632 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -9.70 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.993951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
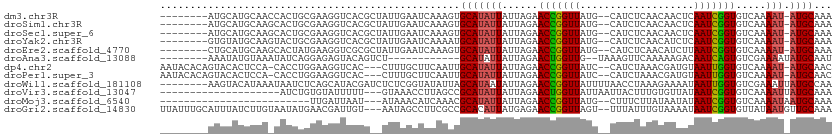
>dm3.chr3R 1424226 101 - 27905053 --------AUGCAUGCAACCACUGCGAAGGUCACGCUAUUGAAUCAAAGUGCAUAUUAUUAGAACCGGUUAUG--CAUCUCAACAACUCAAUCGGUGUCAAAAU-AUGCAAA --------.((((((..(((........))).((((((((((......(((((((..............))))--))).........))))).))))).....)-))))).. ( -18.90, z-score = 0.16, R) >droSim1.chr3R 1460571 101 - 27517382 --------AUGCAUGCAAGCACUGCGAAGGUCACGCUAUUGAAUCAAAGUGCAUAUUAUUAGAACCGGUUAUG--CAUCUCAACAACUCAAUCGGUGUCAAAAU-AUGCAAA --------.((((((...(((((..((...(((......))).))..))))).........(((((((((..(--...........)..))))))).))....)-))))).. ( -21.50, z-score = -0.39, R) >droSec1.super_6 1539288 101 - 4358794 --------AUGCAUGCAAGCACUGCGAAGGUCACGCUAUUGAAUCAAAGUGCAUAUUAUUAGAACCGGUUAUG--CAUCUCAACAACUCAAUCGGUGUCAAAAU-AUGCAAA --------.((((((...(((((..((...(((......))).))..))))).........(((((((((..(--...........)..))))))).))....)-))))).. ( -21.50, z-score = -0.39, R) >droYak2.chr3R 17388921 101 - 28832112 --------GUGUAUGCAAGUACUGCGAAGGUCACGCUAUUGAAUCAAAAUGCAUAUUAUUAGAACCGGUUAUG--CAUCUCAACAUCUCAAUCGGUGUCAAAAU-AUGCAAA --------(((..((((.....)))).....)))...............((((((((......((((((((((--........)))...))))))).....)))-))))).. ( -19.50, z-score = -0.26, R) >droEre2.scaffold_4770 1694643 101 - 17746568 --------CUGCAUGCAAGCACUAUGAAGGUCGCGCUAUUGAAUCAAAGUGCAUAUUAUUAGAACCGGUUAUG--CAUCUCAACAUCUUAAUCGGUGUCAAAAU-AUGCAAA --------.((((((...(((((.(((...(((......))).))).))))).........((((((((((..--.............)))))))).))....)-))))).. ( -24.46, z-score = -1.47, R) >droAna3.scaffold_13088 68097 90 + 569066 --------AAAUAUGUAAAUAUCAGGAGAGUACAGUCU------------GCAUAUUAUUAGAACUGGUUG--UAAAGUUCAAAAAGACAAUCAGUGUCGAAAAUAUGCAAU --------.....((((....((....)).))))...(------------(((((((.((...((((((((--(.............)))))))))...)).)))))))).. ( -20.02, z-score = -3.19, R) >dp4.chr2 7424498 105 + 30794189 AAUACACAGUACACUCCA-CACCUGGAAGGUCAC---CUUUGCUUCAAUUGCAUAUUAUUAGAACCGGUUAUC--CAUCUAAACGAUGUAAUUGGUGUCAAAAU-AUGCAAC ........((((..((((-....))))..)).))---...........(((((((((......((((((((..--((((.....)))))))))))).....)))-)))))). ( -23.30, z-score = -1.89, R) >droPer1.super_3 1234565 105 + 7375914 AAUACACAGUACACUCCA-CACCUGGAAGGUCAC---CUUUGCUUCAAUUGCAUAUUAUUAGAACCGGUUAUC--CAUCUAAACGAUGUAAUUGGUGUCAAAAU-AUGCAAC ........((((..((((-....))))..)).))---...........(((((((((......((((((((..--((((.....)))))))))))).....)))-)))))). ( -23.30, z-score = -1.89, R) >droWil1.scaffold_181108 3226494 104 + 4707319 --------AAGUACAUAAAUAAUCUCAGCAUACGAUCUCUCGGUAUAUUAGCAUAAUAUUAGAACCGGUUAUUUUAACCUAAAGAAAAUAAUUGGUGUCGAAAUUAUGCCAA --------........................(((....)))........(((((((.((...((((((((((((.........))))))))))))...)).)))))))... ( -18.50, z-score = -1.55, R) >droVir3.scaffold_13047 11661588 89 + 19223366 --------------------AUCUGUGUAUUUUU---GUAAACCUUAGCCGCAUAUUAUUAGAACUGGUUAUUAAUUACUUUGUGUUAUAAUCGGUGUCAAAAUUAUGCAAA --------------------...((((((.((((---(...(((.((((((..((....))....)))))).((((........)))).....)))..))))).)))))).. ( -14.20, z-score = -0.46, R) >droMoj3.scaffold_6540 30630577 82 + 34148556 -------------------------UUGAUUAAU---AUAAACAUCAAACGCAUAUUAUUAGAACCGGUUAUG--CUUUCUUAUAAUAUAAUCGGUGUCAAAAUAAUGCAAA -------------------------(((((....---......)))))..((((.........((((((((((--.((......)))))))))))).........))))... ( -14.57, z-score = -2.09, R) >droGri2.scaffold_14830 2852847 107 + 6267026 UUAUUUGCAUUUAUCUUGUAAUAUGAACGAUUGU---AAUAGCCUUCGCCGCACAUUAUGAGAACCGGUUAGU--UUUAUUUGUAAAAUAAUCGGUGUUAUAAUGUUGCAAA (((((((((.......))))).)))).(((..((---....))..)))..(((((((((((..((((((((.(--((((....))))))))))))).)))))))).)))... ( -26.30, z-score = -2.58, R) >consensus ________AUGCAUGCAA_CACUGGGAAGGUCACG__AUUGAAUCAAAGUGCAUAUUAUUAGAACCGGUUAUG__CAUCUCAACAACAUAAUCGGUGUCAAAAU_AUGCAAA ..................................................((((.........(((((((...................))))))).........))))... ( -9.70 = -9.38 + -0.32)
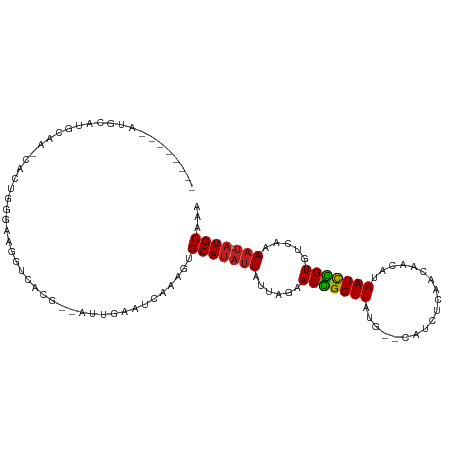
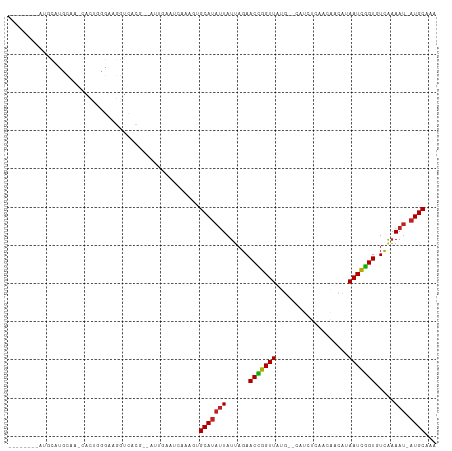
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:45 2011