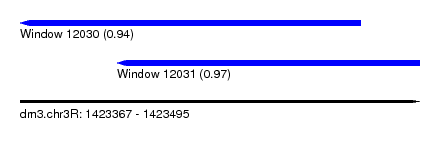
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,423,367 – 1,423,495 |
| Length | 128 |
| Max. P | 0.970629 |

| Location | 1,423,367 – 1,423,476 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.63911 |
| G+C content | 0.32219 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -11.53 |
| Energy contribution | -11.24 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1423367 109 - 27905053 ----AUUUGUUUUUAGCCCCGGAAUGUUCUCAGGAAGAUG-------ACUCAAUGAUCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGAGUAAGAAAGUUCGACGGAGUUUUACCCAAUU ----..(((..(..(((.((((((((((.....(((((((-------.(.((((......)))).))))))))....))))))))..(((........)))...)).)))..)..))).. ( -24.10, z-score = -1.13, R) >droGri2.scaffold_14830 2851923 109 + 6267026 CCUCAUAUCAAUAUCA-UUCGGAAUGUUUUCAGGAAGAU--------CUACAUGCAUAUC-UUGUUAAUCUUUAAUUAAUAUUCCUAGA-GUAAUUAUUUAUGCUGAAUUUUAUGCAUGU ...............(-(((((((((((.....((((((--------..(((........-.)))..))))))....))))))))..))-)).......(((((..........))))). ( -19.30, z-score = -0.94, R) >droMoj3.scaffold_6540 30629516 106 + 34148556 ----AGAGGCACAUAC-UUUGGAAUGUUUUCAGGAAGAU--------CUACAUGUAUAUA-UUGUUAAUCUUUAAUUAACAUUCCUAGAUAAAAGCAUUUACAUGGAAUUUAAUGCAUUU ----((((.......)-)))((((((((.....((((((--------..(((........-.)))..))))))....)))))))).........(((((............))))).... ( -21.10, z-score = -1.93, R) >droVir3.scaffold_13047 11660742 104 + 19223366 ----AUUUGUAAAUA--UUUGGAAUGUUUUCAGGAAGAU--------CUACAUGCAUAUC-UUGUUAAUCUUUAAUUAACAUUCCUAGA-GCAAUCAUUUACAUGGAAUUUAAUGCAUGU ----...(((((((.--...((((((((.....((((((--------..(((........-.)))..))))))....))))))))..((-....)))))))))................. ( -22.90, z-score = -2.24, R) >droWil1.scaffold_181108 3225608 108 + 4707319 ----UCUUGAUUUUGG--UCGGAAAGUUAUCAGGAAGAUGUUGCAACAUUUAUUAAUCCAGUUGUACAUCUUUAAUUGACGUUCCUAGAAUA------UAGUUCGGAAUUUUACACUAGU ----(((.((((....--((((((.((((....((((((((.(((((.............)))))))))))))...)))).))))..))...------.)))).)))............. ( -23.52, z-score = -1.47, R) >droPer1.super_3 1233731 105 + 7375914 ----GACUGCAGUCUGUUAUGGAAUGUUCUCAGGAAGAUG-------UUAUAACCAUUCAGUUGUGUAUCUUUAAUUGAUGUUCCUAGAGUAAA----UUUCGUGGAGUUUUAUUUAUUU ----(((....)))..(((((((((...((((((((....-------.((((((......))))))((((.......))))))))).)))...)----)))))))).............. ( -21.30, z-score = -0.88, R) >dp4.chr2 7423664 105 + 30794189 ----GACUGUAGUCUCUUAUGGAAUGUUCUCAGGAAGAUG-------UUAUAACCAUUCAGUUGUGUAUCUUUAAUUGAUGUUCCUAGACUAAA----UUUCGUGGAGUUUUAUUUAUUU ----((...((((((.....((((((((.....(((((((-------(.(((((......)))))))))))))....)))))))).))))))..----..)).................. ( -20.20, z-score = -0.56, R) >droAna3.scaffold_13088 67249 107 + 569066 ----UUUUAUAUUUAUGUCUGGAAUAUUUUCAGGAAGAUG-------AUUCAAUCGUUUAGUUGUACAUCUUUAAUAAUUAUUCCUAGA-UAACAAA-UUCGUCGGAGUAUUACAUAUUU ----...........(((((((((((.((....(((((((-------...((((......))))..)))))))...)).)))))).)))-))....(-(((....))))........... ( -20.60, z-score = -1.79, R) >droEre2.scaffold_4770 1693826 108 - 17746568 ----AUUUGUUUCUAG-CCCGGAAUGUUCUCAGGAAGAUG-------UCUCGAUCACCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGAGUAAGAAAGUUCGUCGGAUUUCUACCCAAUU ----.....(((((.(-((.((((((((.....(((((((-------(..((((......)))).))))))))....))))))))..).)).))))).......((........)).... ( -23.20, z-score = -0.78, R) >droYak2.chr3R 17388077 108 - 28832112 ----GUUUGUUUUCAG-CCCGGAAUGUUCUCAGGAAGAUG-------ACUCAAUCAUUCAGUUGUACAUCUUUAAUUGAUAUUCCUAGAGUAAGAAAAUCCGACGGAUUUUUACCCAAUU ----............-...((((((((.....(((((((-------...((((......))))..)))))))....))))))))........((((((((...))))))))........ ( -22.80, z-score = -1.15, R) >droSec1.super_6 1538432 109 - 4358794 ----AUUUGUUUUUAGCCCCGGAAUGUUCUCAGGAAGAUG-------ACUCAAUGAUCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGAGUAAGAAAGUUCGACGGAGUUUUACCCAUUU ----...((..(..(((.((((((((((.....(((((((-------.(.((((......)))).))))))))....))))))))..(((........)))...)).)))..)..))... ( -23.20, z-score = -0.83, R) >consensus ____AUUUGUAUUUAG_UCCGGAAUGUUCUCAGGAAGAUG_______AUUCAAUCAUCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGAGUAAAAAA_UUCGACGGAGUUUUACCCAUUU ....................((((((((.....(((((((..........................)))))))....))))))))................................... (-11.53 = -11.24 + -0.29)

| Location | 1,423,398 – 1,423,495 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.40 |
| Shannon entropy | 0.63106 |
| G+C content | 0.30902 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -11.53 |
| Energy contribution | -11.24 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
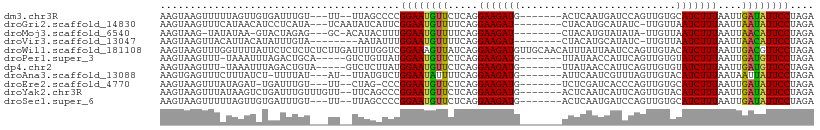
>dm3.chr3R 1423398 97 - 27905053 AAGUAAGUUUUUAGUUGUGAUUUGU---UU--UUAGCCCCGGAAUGUUCUCAGGAAGAUG-------ACUCAAUGAUCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGA .............((((.((.....---))--.))))...((((((((.....(((((((-------.(.((((......)))).))))))))....)))))))).... ( -19.50, z-score = -0.95, R) >droGri2.scaffold_14830 2851953 97 + 6267026 AAGUAAGUUUCAUAACAUCCUCAUA---UCAAUAUCAUUCGGAAUGUUUUCAGGAAGAU--------CUACAUGCAUAUC-UUGUUAAUCUUUAAUUAAUAUUCCUAGA ......(((....))).........---............((((((((.....((((((--------..(((........-.)))..))))))....)))))))).... ( -15.50, z-score = -1.55, R) >droMoj3.scaffold_6540 30629547 94 + 34148556 AAGUAAG-UAUAUAA-GUACUAGAG---GC-ACAUACUUUGGAAUGUUUUCAGGAAGAU--------CUACAUGUAUAUA-UUGUUAAUCUUUAAUUAACAUUCCUAGA .....((-(((....-)))))((((---..-.....))))((((((((.....((((((--------..(((........-.)))..))))))....)))))))).... ( -20.20, z-score = -1.78, R) >droVir3.scaffold_13047 11660772 92 + 19223366 AAGUAAGUUACAUUACAUAUUUGUA--------AAUAUUUGGAAUGUUUUCAGGAAGAU--------CUACAUGCAUAUC-UUGUUAAUCUUUAAUUAACAUUCCUAGA ............(((((....))))--------)......((((((((.....((((((--------..(((........-.)))..))))))....)))))))).... ( -19.50, z-score = -2.58, R) >droWil1.scaffold_181108 3225633 109 + 4707319 AAGUAAGUUUGGUUUUAUUCUCUCUCUCUUGAUUUUGGUCGGAAAGUUAUCAGGAAGAUGUUGCAACAUUUAUUAAUCCAGUUGUACAUCUUUAAUUGACGUUCCUAGA (((..((...((......))...))..)))(((....)))((((.((((....((((((((.(((((.............)))))))))))))...)))).)))).... ( -22.62, z-score = -1.01, R) >droPer1.super_3 1233758 96 + 7375914 AAGUAAGUUU-UAAAUUUAGACUGCA-----GUCUGUUAUGGAAUGUUCUCAGGAAGAUG-------UUAUAACCAUUCAGUUGUGUAUCUUUAAUUGAUGUUCCUAGA ..........-......(((((....-----)))))....((((((((.....(((((((-------(.(((((......)))))))))))))....)))))))).... ( -18.30, z-score = -0.68, R) >dp4.chr2 7423691 96 + 30794189 AAGUAAGUUU-UAAAUUUAGACUGUA-----GUCUCUUAUGGAAUGUUCUCAGGAAGAUG-------UUAUAACCAUUCAGUUGUGUAUCUUUAAUUGAUGUUCCUAGA ..(((((...-.......((((....-----)))))))))((((((((.....(((((((-------(.(((((......)))))))))))))....)))))))).... ( -17.80, z-score = -0.64, R) >droAna3.scaffold_13088 67278 96 + 569066 AAGUGAGUUUCUUUAUCU-UUUUAU---AU--UUAUGUCUGGAAUAUUUUCAGGAAGAUG-------AUUCAAUCGUUUAGUUGUACAUCUUUAAUAAUUAUUCCUAGA ..................-......---..--.....(((((((((.((....(((((((-------...((((......))))..)))))))...)).)))))).))) ( -16.40, z-score = -0.87, R) >droEre2.scaffold_4770 1693857 95 - 17746568 AAGUAAGUUUAUAGAU-UGAUUUGU---UU--CUAG-CCCGGAAUGUUCUCAGGAAGAUG-------UCUCGAUCACCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGA ..((.((...(((((.-...)))))---..--)).)-)..((((((((.....(((((((-------(..((((......)))).))))))))....)))))))).... ( -20.20, z-score = -1.02, R) >droYak2.chr3R 17388108 100 - 28832112 AAGUAAGUUUAUAAGUCUGAUUUGUUUGUU--UUCAGCCCGGAAUGUUCUCAGGAAGAUG-------ACUCAAUCAUUCAGUUGUACAUCUUUAAUUGAUAUUCCUAGA ..............(.((((..........--.)))))..((((((((.....(((((((-------...((((......))))..)))))))....)))))))).... ( -18.90, z-score = -0.45, R) >droSec1.super_6 1538463 97 - 4358794 AAGUAAGUUUUUAGUUGUGAUUUGU---UU--UUAGCCCCGGAAUGUUCUCAGGAAGAUG-------ACUCAAUGAUCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGA .............((((.((.....---))--.))))...((((((((.....(((((((-------.(.((((......)))).))))))))....)))))))).... ( -19.50, z-score = -0.95, R) >consensus AAGUAAGUUUCUAAAUAUAAUUUGU___UU__UUAGCUCCGGAAUGUUCUCAGGAAGAUG_______AUUCAAUCAUCCAGUUGUGCAUCUUUAAUUGAUAUUCCUAGA ........................................((((((((.....(((((((..........................)))))))....)))))))).... (-11.53 = -11.24 + -0.29)
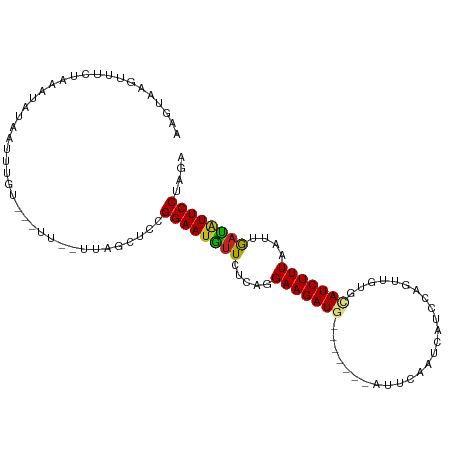
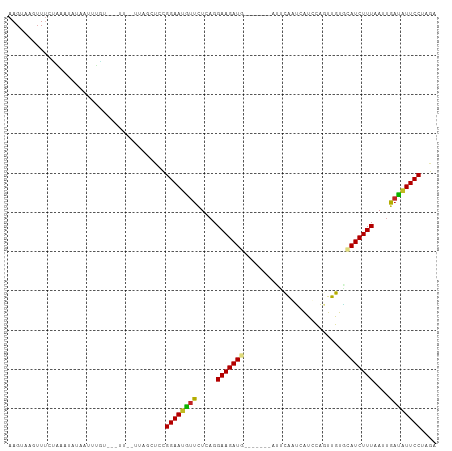
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:42 2011