| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,422,237 – 1,422,317 |
| Length | 80 |
| Max. P | 0.999586 |

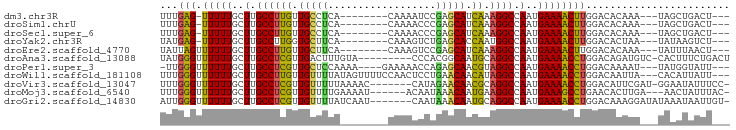
| Location | 1,422,237 – 1,422,317 |
|---|---|
| Length | 80 |
| Sequences | 11 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 67.88 |
| Shannon entropy | 0.64773 |
| G+C content | 0.39219 |
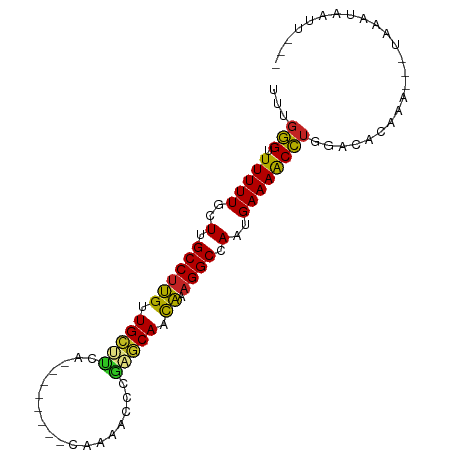
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -13.07 |
| Energy contribution | -11.72 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1422237 80 - 27905053 UUUGAG-UUUUUGCUUGCCUUGUUGCCUCA--------CAAAAUCCGAGCAUCAAAGGCCAAUGAAAACUUGGACACAAA---UAGCUGACU--- ((..((-(((((..(.(((((..(((.((.--------........)))))...))))).)..)))))))..))......---.........--- ( -19.40, z-score = -1.57, R) >droSim1.chrU 11235156 80 - 15797150 UUUGAG-UUUUUGCUUGCCUUGUUGCCUCA--------CAAAACCCGAGCAUCAAAGGCCAAUGAAAACUUGGACACAAA---UAGCUGACU--- ((..((-(((((..(.(((((..(((.((.--------........)))))...))))).)..)))))))..))......---.........--- ( -19.40, z-score = -1.69, R) >droSec1.super_6 1537302 80 - 4358794 UUUGAG-UUUUUGCUUGCCUUGUUGCCUCA--------CAAAACCCGAGCAUCAAAGGCCAAUGAAAACUUGGACACAAA---UAGCUGACU--- ((..((-(((((..(.(((((..(((.((.--------........)))))...))))).)..)))))))..))......---.........--- ( -19.40, z-score = -1.69, R) >droYak2.chr3R 17386936 80 - 28832112 UAUGAG-UUUUUGCUUGCCUUGGUGCUUCA--------CAAAGUCUGAGCACCAAUGGCCAAUGAAAACUUGGACACUAA---UAUAAGUCU--- ...(((-(((((..(.(((((((((((.((--------.......)))))))))).))).)..))))))))((((.....---.....))))--- ( -25.30, z-score = -3.34, R) >droEre2.scaffold_4770 1692722 81 - 17746568 UAUUAGUUUUUUGCUUGCCUUGUUGCUUCA--------CAAAGUCCGAGCAUCAAAGGCCAAUGAAAACUUGGACACAAA---UAUUUAACU--- ....(((((((.....(((((..(((((..--------........)))))...)))))....)))))))..........---.........--- ( -15.50, z-score = -0.91, R) >droAna3.scaffold_13088 66278 85 + 569066 UAUGGGUUUUUUGCUUGCCUCGUUGACUUUGUA---------CCCACGGCAAUGCAGGCCAAUGAAAACCUGGACAGAUGUC-CACUUUCUGACU ...((((((((.....(((((((((.((.((..---------..)).))))))).))))....))))))))((((....)))-)........... ( -25.60, z-score = -2.22, R) >droPer1.super_3 1232552 84 + 7375914 -UUGGGUUUUUUGCUUGCCUCGUUGCUCCAAAA----GAAAAACCAGAGCAACGUAGGCCAAUGAAAACCUGGACAAAAU---UAUGGUAUU--- -(..(((((((.....((((((((((((.....----.........)))))))).))))....)))))))..).......---.........--- ( -27.44, z-score = -3.40, R) >droWil1.scaffold_181108 3224631 89 + 4707319 UUUGGGUUUUUUGCUUGCCUUGUUGUUUUAUAGUUUUCCAACUCCUGAACAACAUAGGCCAAUGAAAACCUGGACAAUUA---CACAUUAUU--- ((..(((((((.....((((((((((((...((((....))))...)))))))).))))....)))))))..))......---.........--- ( -26.00, z-score = -4.03, R) >droVir3.scaffold_13047 11659767 86 + 19223366 UUUGGGUUUUUUGCUUGCCUCGUUGUUUUUAAAAC-------CAUAGAACAACGCAGGCCAAUGAAAACCUGGACAUUCGAU-GGAAUAUUUCC- ((..(((((((.....(((((((((((((......-------...))))))))).))))....)))))))..)).((((...-.))))......- ( -26.70, z-score = -3.45, R) >droMoj3.scaffold_6540 30628486 85 + 34148556 UUUGGGUUUUUUGCUUGCCUCGUUGUUUUGAAAAU------ACAAUAAACAAUGAAGGCCAAUGAAAGCCUGAACACUUGA---AACUAUUUAC- ((..(((((((.....((((((((((((.......------.....)))))))).))))....)))))))..)).......---..........- ( -22.30, z-score = -3.51, R) >droGri2.scaffold_14830 2850955 87 + 6267026 AUUGGGUUUUUUGCUUGCCUCGUUGUUUUAUCAAU-------CAAUAAACAAUGCAGGCCAAUGAAAACCUGGACAAAGGAUAUAAAUAAUUGU- .(..(((((((.....((((((((((((.......-------....)))))))).))))....)))))))..).....................- ( -22.10, z-score = -2.24, R) >consensus UUUGGGUUUUUUGCUUGCCUUGUUGCUUCA________CAAAACCCGAGCAACAAAGGCCAAUGAAAACCUGGACACAAA___UAAAUAAUU___ ...(((.(((((..(.((((((.(((((..................))))).)).)))).)..))))))))........................ (-13.07 = -11.72 + -1.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:41 2011