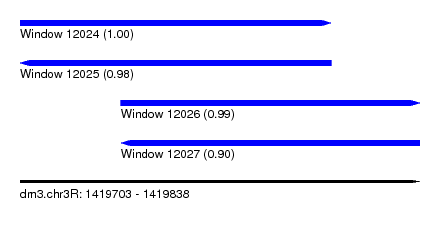
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,419,703 – 1,419,838 |
| Length | 135 |
| Max. P | 0.999132 |

| Location | 1,419,703 – 1,419,808 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 64.44 |
| Shannon entropy | 0.73096 |
| G+C content | 0.52413 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -12.99 |
| Energy contribution | -11.78 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
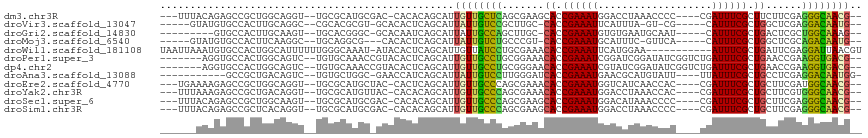
>dm3.chr3R 1419703 105 + 27905053 ---UUUACAGAGCCGCUGGCAGGU--UGCGCAUGCGAC-CACACAGCAUUGUUGCUCAGCGAAGCACCGAAAUGGACCUAAACCCC----CGAUUUCGCUUCUUCGAGGGCAACG-- ---...........((((...(((--(((....)))))-)...))))...(((((((..(((((...((((((((..........)----).))))))...))))).))))))).-- ( -37.70, z-score = -2.77, R) >droVir3.scaffold_13047 11657234 99 - 19223366 -----GUAUGUGCCACUUGCAGGC--CGCACGCGU-GCACACUCAGCAUUAUUGUCCGCUUGC-CACCGAAAUUCAUUUA-GU-CG-----CAUUUCGCUGGCUCGAGGACAAUG-- -----(..((((((.(.(((....--.))).).).-)))))..).....((((((((.(..((-((.((((((.(.....-..-.)-----.)))))).))))..).))))))))-- ( -33.30, z-score = -1.94, R) >droGri2.scaffold_14830 2848144 97 - 6267026 ---------GUGCCACUUGCAAGU--UGCACGGGC-GCACAAUCAGCAUUAUUGCCAGCUUGC-CACCGAAAUGUGUGAAUGCAAU-----CAUUUCGCUGACUCGCUGGCAAAG-- ---------(((((.(.(((....--.))).))))-)).............((((((((..(.-((.((((((((((....)))..-----))))))).)).)..))))))))..-- ( -36.60, z-score = -2.44, R) >droMoj3.scaffold_6540 30624310 98 - 34148556 -----GUAUGUGCCACUUCAAGGC--UGCAGGCG---CACACUCAGCAUUAUUGUCUGCCCGU-CACCGAAAUGCAUUUC-GUUCA-----CAUUUCGCUGGCUCGCAGACAAUG-- -----((.((((((..........--....))))---)).)).......((((((((((..((-((.(((((((......-.....-----))))))).))))..))))))))))-- ( -33.84, z-score = -2.98, R) >droWil1.scaffold_181108 3221552 105 - 4707319 UAAUUAAAUGUGCCACUGGCAUUUUUUGGGCAAAU-AUACACUCAGCAUUGUUAUCCUGCGAAACACCGAAAUUCAUGGAA-----------AUUUCGCUGAUUCGAGGAUUAACGU ......((((((((...)))......((((.....-.....)))))))))((((((((.((((.((.(((((((......)-----------)))))).)).)))))))).)))).. ( -25.30, z-score = -1.72, R) >droPer1.super_3 1229233 106 - 7375914 -------AGGUGCCACUGGCAGUC--UGUGCAAACCGUACACUCAGCAUUGUUGCCUGCGGAAACACCGAAAUCGGAUCGGAUAUCGGUCUGAUUUCGCUGAACCGAAGGUGACG-- -------...((((...))))(..--(((((.....)))))..)......((..(((.(((...((.(((((((((((((.....))))))))))))).))..))).)))..)).-- ( -41.80, z-score = -3.10, R) >dp4.chr2 7418833 106 - 30794189 -------AGGUGCCACUGGCAGUC--UGUGCAAACCGUACACUCAGCAUUGUUGCCUGCGGGAACACCGAAAUCGUAUCGGAUAUCGGUCUGAUUUCGCUGAACCGAAGGUGACG-- -------...((((...))))(..--(((((.....)))))..)......((..(((.(((...((.((((((((.((((.....)))).)))))))).))..))).)))..)).-- ( -36.70, z-score = -1.51, R) >droAna3.scaffold_13088 63427 98 - 569066 -----------GCCGCUGACAGUC--UGUGCUGGC-GAACCAUCAGCAUUAUUGUCCUUGGGAUCACCGAAAUGAACGCAUGUAUU----UUAUUUCGCUGCCUCGAGGACAAUGG- -----------...(((((..((.--(((....))-).))..)))))..(((((((((((((..((.(((((((((.........)----)))))))).)).))))))))))))).- ( -34.80, z-score = -3.06, R) >droEre2.scaffold_4770 1689790 105 + 17746568 ---UGAAAAGAGCCGCUGGCAGGU--UGCGCAUGCUAC-CACUCAGCAUUGUUGCCCAGCGAAACACCGAAAUGGUCAUCAACCAC----CGAUUUCGCUGCUUCGAUGGCAACG-- ---...........(((((..(((--.((....)).))-)..)))))...((((((...((((.((.(((((((((........))----).)))))).)).))))..)))))).-- ( -36.10, z-score = -2.25, R) >droYak2.chr3R 17383611 105 + 28832112 ---UUUAAAGAGCCGCUGACAGGU--UGCGCAUGUUAC-CACACAGCAUUGUUGCCCAGCGAAACACCGAAAUGGACCUAAACCAC----CGAUUUCGCUGCUUCGUGGGCAACG-- ---...........((((...(((--.((....)).))-)...))))...((((((((.((((.((.((((((((..........)----).)))))).)).)))))))))))).-- ( -36.30, z-score = -3.34, R) >droSec1.super_6 1534588 105 + 4358794 ---UUUACAGAGCCGCUGGCAAGU--UGCGCAUGCGAC-CACACAGCAUUGUUGCCCAGCGAAGCACCGAAAUGGACAUAAACCCC----CGAUUUCGCUGCUUCGAGGGCAACG-- ---...........((((....((--(((....)))))-....))))...(((((((..(((((((.((((((((..........)----).)))))).))))))).))))))).-- ( -43.50, z-score = -4.93, R) >droSim1.chr3R 1456141 105 + 27517382 ---UUUACAGAGCCGCUCACAGGU--UGCGCAUGCGAC-CACACAGCAUUGUUGCCCAGCGAAGCACCGAAAUGGACCUAAACCCC----CGAUUUCGCUGCUUCGAGGGCAACG-- ---...........(((....(((--(((....)))))-)....)))...(((((((..(((((((.((((((((..........)----).)))))).))))))).))))))).-- ( -42.90, z-score = -5.22, R) >consensus ______AAAGUGCCACUGGCAGGU__UGCGCAUGC_ACACACUCAGCAUUGUUGCCCAGCGAAACACCGAAAUGGACUUAAACCCC____CGAUUUCGCUGCUUCGAGGGCAACG__ ..................................................(((((((.......((.((((((...................)))))).))......)))))))... (-12.99 = -11.78 + -1.21)
| Location | 1,419,703 – 1,419,808 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.44 |
| Shannon entropy | 0.73096 |
| G+C content | 0.52413 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -12.04 |
| Energy contribution | -10.45 |
| Covariance contribution | -1.59 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1419703 105 - 27905053 --CGUUGCCCUCGAAGAAGCGAAAUCG----GGGGUUUAGGUCCAUUUCGGUGCUUCGCUGAGCAACAAUGCUGUGUG-GUCGCAUGCGCA--ACCUGCCAGCGGCUCUGUAAA--- --.((((((..(((((.(.((((((.(----((........))))))))).).)))))..).)))))..(((((.(((-((.((....)).--))).)))))))..........--- ( -35.00, z-score = 0.09, R) >droVir3.scaffold_13047 11657234 99 + 19223366 --CAUUGUCCUCGAGCCAGCGAAAUG-----CG-AC-UAAAUGAAUUUCGGUG-GCAAGCGGACAAUAAUGCUGAGUGUGC-ACGCGUGCG--GCCUGCAAGUGGCACAUAC----- --.(((((((.(..((((.((((((.-----..-..-.......)))))).))-))..).)))))))........((((((-.(((.(((.--....))).)))))))))..----- ( -39.40, z-score = -2.68, R) >droGri2.scaffold_14830 2848144 97 + 6267026 --CUUUGCCAGCGAGUCAGCGAAAUG-----AUUGCAUUCACACAUUUCGGUG-GCAAGCUGGCAAUAAUGCUGAUUGUGC-GCCCGUGCA--ACUUGCAAGUGGCAC--------- --..((((((((..((((.(((((((-----..((....))..))))))).))-))..))))))))....((.......))-((((.(((.--....))).).)))..--------- ( -38.80, z-score = -2.36, R) >droMoj3.scaffold_6540 30624310 98 + 34148556 --CAUUGUCUGCGAGCCAGCGAAAUG-----UGAAC-GAAAUGCAUUUCGGUG-ACGGGCAGACAAUAAUGCUGAGUGUG---CGCCUGCA--GCCUUGAAGUGGCACAUAC----- --.(((((((((..(.((.(((((((-----(....-.....)))))))).))-.)..)))))))))........(((((---.(((.((.--........))))).)))))----- ( -35.30, z-score = -2.02, R) >droWil1.scaffold_181108 3221552 105 + 4707319 ACGUUAAUCCUCGAAUCAGCGAAAU-----------UUCCAUGAAUUUCGGUGUUUCGCAGGAUAACAAUGCUGAGUGUAU-AUUUGCCCAAAAAAUGCCAGUGGCACAUUUAAUUA ..((((.((((((((.((.((((((-----------((....)))))))).)).)))).)))))))).....(((((((.(-(((.((.........)).))))..))))))).... ( -28.80, z-score = -2.75, R) >droPer1.super_3 1229233 106 + 7375914 --CGUCACCUUCGGUUCAGCGAAAUCAGACCGAUAUCCGAUCCGAUUUCGGUGUUUCCGCAGGCAACAAUGCUGAGUGUACGGUUUGCACA--GACUGCCAGUGGCACCU------- --.(((((....((..((.(((((((.((.((.....)).)).))))))).))...))(((((((....)))...(((((.....))))).--..))))..)))))....------- ( -35.10, z-score = -1.55, R) >dp4.chr2 7418833 106 + 30794189 --CGUCACCUUCGGUUCAGCGAAAUCAGACCGAUAUCCGAUACGAUUUCGGUGUUCCCGCAGGCAACAAUGCUGAGUGUACGGUUUGCACA--GACUGCCAGUGGCACCU------- --.(((((....((..((.(((((((....((.....))....))))))).))..)).(((((((....)))...(((((.....))))).--..))))..)))))....------- ( -32.40, z-score = -0.80, R) >droAna3.scaffold_13088 63427 98 + 569066 -CCAUUGUCCUCGAGGCAGCGAAAUAA----AAUACAUGCGUUCAUUUCGGUGAUCCCAAGGACAAUAAUGCUGAUGGUUC-GCCAGCACA--GACUGUCAGCGGC----------- -((((((((((.(.((((.((((((..----...((....))..)))))).))..))).))))))))...(((((((((..-(......).--.))))))))))).----------- ( -32.30, z-score = -2.31, R) >droEre2.scaffold_4770 1689790 105 - 17746568 --CGUUGCCAUCGAAGCAGCGAAAUCG----GUGGUUGAUGACCAUUUCGGUGUUUCGCUGGGCAACAAUGCUGAGUG-GUAGCAUGCGCA--ACCUGCCAGCGGCUCUUUUCA--- --.((((((..(((((((.((((((.(----((........))))))))).)))))))...))))))..(((((.(((-((.((....)).--))).)))))))..........--- ( -44.10, z-score = -2.73, R) >droYak2.chr3R 17383611 105 - 28832112 --CGUUGCCCACGAAGCAGCGAAAUCG----GUGGUUUAGGUCCAUUUCGGUGUUUCGCUGGGCAACAAUGCUGUGUG-GUAACAUGCGCA--ACCUGUCAGCGGCUCUUUAAA--- --.(((((((((((((((.((((((.(----(..........)))))))).))))))).))))))))..(((((...(-((..........--)))...)))))..........--- ( -42.40, z-score = -3.11, R) >droSec1.super_6 1534588 105 - 4358794 --CGUUGCCCUCGAAGCAGCGAAAUCG----GGGGUUUAUGUCCAUUUCGGUGCUUCGCUGGGCAACAAUGCUGUGUG-GUCGCAUGCGCA--ACUUGCCAGCGGCUCUGUAAA--- --.(((((((.(((((((.((((((.(----((.(....).))))))))).)))))))..)))))))..(((...(.(-(((((.((.((.--....))))))))))).)))..--- ( -47.80, z-score = -3.64, R) >droSim1.chr3R 1456141 105 - 27517382 --CGUUGCCCUCGAAGCAGCGAAAUCG----GGGGUUUAGGUCCAUUUCGGUGCUUCGCUGGGCAACAAUGCUGUGUG-GUCGCAUGCGCA--ACCUGUGAGCGGCUCUGUAAA--- --.(((((((.(((((((.((((((.(----((........))))))))).)))))))..)))))))...(((((..(-((.((....)).--))).....)))))........--- ( -47.20, z-score = -3.28, R) >consensus __CGUUGCCCUCGAAGCAGCGAAAUCG____GGGGUUUAAGUCCAUUUCGGUGUUUCGCCGGGCAACAAUGCUGAGUGUGU_GCAUGCGCA__ACCUGCCAGCGGCACUUU______ ...((.((((......((.((((((...................)))))).)).......)))).))...((((.(((.....)))(((.......)))...))))........... (-12.04 = -10.45 + -1.59)

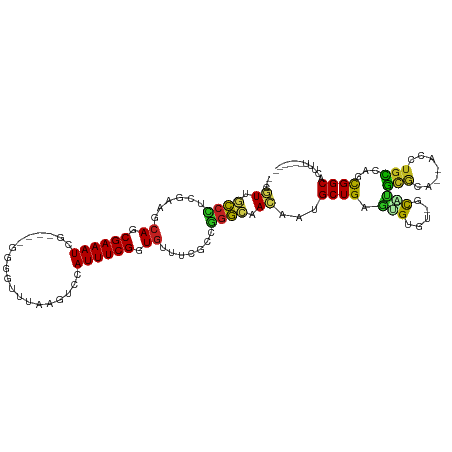
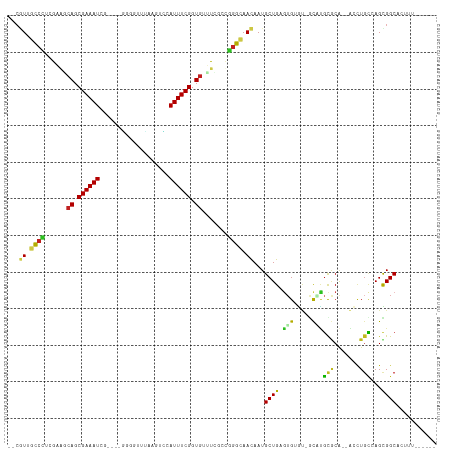
| Location | 1,419,737 – 1,419,838 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.36 |
| Shannon entropy | 0.68088 |
| G+C content | 0.52490 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -11.87 |
| Energy contribution | -10.56 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1419737 101 + 27905053 ACACAGCAUUGUUGCUCAGCGAAGCACCGAAAUGGACCUAAACCCC----CGAUUUCGCUUCUUCGAGGGCAA---CGGAC-GCUUGUGCAACUGCCACUGGCUCAACG-------- .((((((.(((((((((..(((((...((((((((..........)----).))))))...))))).))))))---)))..-)).)))).....(((...)))......-------- ( -30.30, z-score = -1.52, R) >droVir3.scaffold_13047 11657266 98 - 19223366 ACUCAGCAUUAUUGUCCGCUUG-CCACCGAAAUUCAUUUA-GU--C----GCAUUUCGCUGGCUCGAGGACAA---UGUGC-GCCGGCGUCCA-GCAACUGGCACAUUUUA------ .....((((.(((((((.(..(-(((.((((((.(.....-..--.----).)))))).))))..).))))))---)))))-(((((......-....)))))........------ ( -34.60, z-score = -3.10, R) >droGri2.scaffold_14830 2848172 107 - 6267026 AAUCAGCAUUAUUGCCAGCUUG-CCACCGAAAUGUGUGAAUGCA-A----UCAUUUCGCUGACUCGCUGGCAA---AGUGC-GUCUGUGCACUUGCAACUGGCACAUUGUUGUUAUA ...(((((...((((((((..(-.((.((((((((((....)))-.----.))))))).)).)..))))))))---.((((-.....(((....)))....))))..)))))..... ( -36.70, z-score = -2.16, R) >droMoj3.scaffold_6540 30624340 100 - 34148556 ACUCAGCAUUAUUGUCUGCCCG-UCACCGAAAUGCAUUUC-GUU-C----ACAUUUCGCUGGCUCGCAGACAA---UGUGC-GCCUGCGUCCUUGCAACUGGCACAUGUUG------ .....((((.(((((((((..(-(((.(((((((......-...-.----.))))))).))))..))))))))---)))))-(((((((....))))...)))........------ ( -37.50, z-score = -4.03, R) >droWil1.scaffold_181108 3221591 96 - 4707319 ACUCAGCAUUGUUAUCCUGCGAAACACCGAAAUU-CAUGGAA----------AUUUCGCUGAUUCGAGGAUUAACGUAUAC-AUUUGCCCAAUUGCCACUGGCACAAG--------- .....(((.(((((((((.((((.((.(((((((-......)----------)))))).)).)))))))).))))).....-...))).....((((...))))....--------- ( -22.90, z-score = -2.00, R) >droPer1.super_3 1229264 104 - 7375914 ACUCAGCAUUGUUGCCUGCGGAAACACCGAAAUCGGAUCGGAUAUCGGUCUGAUUUCGCUGAACCGAAGGUGA---CGUAC-GAUUGCGCAGCAGCCACUGGCACAGG--------- .....((...((..(((.(((...((.(((((((((((((.....))))))))))))).))..))).)))..)---)(((.-...)))))....(((...))).....--------- ( -39.10, z-score = -2.56, R) >dp4.chr2 7418864 104 - 30794189 ACUCAGCAUUGUUGCCUGCGGGAACACCGAAAUCGUAUCGGAUAUCGGUCUGAUUUCGCUGAACCGAAGGUGA---CGUAC-GAUUGCGCAGCAGCCACUGGCACAGG--------- .....((...((..(((.(((...((.((((((((.((((.....)))).)))))))).))..))).)))..)---)(((.-...)))))....(((...))).....--------- ( -34.00, z-score = -0.89, R) >droAna3.scaffold_13088 63453 103 - 569066 CAUCAGCAUUAUUGUCCUUGGGAUCACCGAAAUGAACGCAUGUAUU----UUAUUUCGCUGCCUCGAGGACAA---UGGGGAGCCUGUGCAGUUGUCACCGGCACAUACA------- ..((....((((((((((((((..((.(((((((((.........)----)))))))).)).)))))))))))---))).))(((.((((....).))).))).......------- ( -34.90, z-score = -2.33, R) >droEre2.scaffold_4770 1689824 101 + 17746568 ACUCAGCAUUGUUGCCCAGCGAAACACCGAAAUGGUCAUCAACCAC----CGAUUUCGCUGCUUCGAUGGCAA---CGGAC-GCGUGUGCAGCUGCCACUGGCUCAACA-------- ....(((.((((((((...((((.((.(((((((((........))----).)))))).)).))))..)))))---)))..-..(((.((....)))))..))).....-------- ( -32.20, z-score = -1.00, R) >droYak2.chr3R 17383645 101 + 28832112 ACACAGCAUUGUUGCCCAGCGAAACACCGAAAUGGACCUAAACCAC----CGAUUUCGCUGCUUCGUGGGCAA---CGGAC-GCAUGUGCAGCUGCCACCGGCUCAACA-------- .((((((.((((((((((.((((.((.((((((((..........)----).)))))).)).)))))))))))---)))..-)).)))).(((((....))))).....-------- ( -37.10, z-score = -2.89, R) >droSec1.super_6 1534622 101 + 4358794 ACACAGCAUUGUUGCCCAGCGAAGCACCGAAAUGGACAUAAACCCC----CGAUUUCGCUGCUUCGAGGGCAA---CGGAC-GCUUGUGCAACUGUCACUGGCUCAACG-------- .........((((((((..(((((((.((((((((..........)----).)))))).))))))).))))))---))((.-(((.((((....).))).)))))....-------- ( -38.40, z-score = -3.65, R) >droSim1.chr3R 1456175 101 + 27517382 ACACAGCAUUGUUGCCCAGCGAAGCACCGAAAUGGACCUAAACCCC----CGAUUUCGCUGCUUCGAGGGCAA---CGGAC-GCGUGUGCAACUGUCACUGGCUCAACG-------- ....(((..((((((((..(((((((.((((((((..........)----).)))))).))))))).))))))---))(((-(.((.....))))))....))).....-------- ( -38.00, z-score = -2.96, R) >consensus ACUCAGCAUUGUUGCCCAGCGAAACACCGAAAUGGACUUAAACC_C____CGAUUUCGCUGCUUCGAGGGCAA___CGGAC_GCUUGUGCAGCUGCCACUGGCACAACG________ .....((....((((((.......((.((((((...................)))))).))......))))))...............((....)).....)).............. (-11.87 = -10.56 + -1.31)

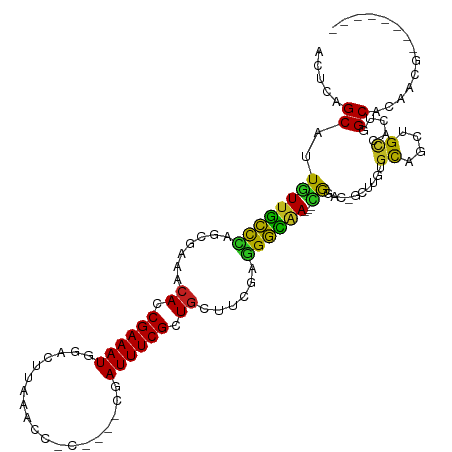
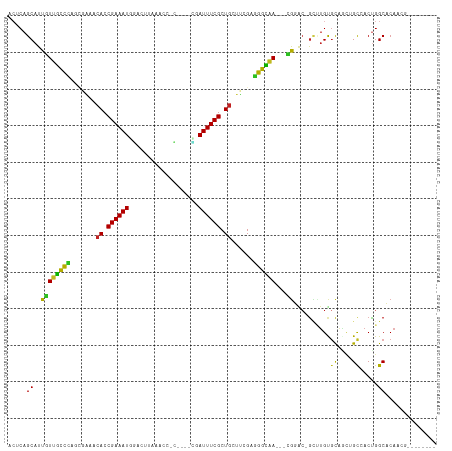
| Location | 1,419,737 – 1,419,838 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.36 |
| Shannon entropy | 0.68088 |
| G+C content | 0.52490 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -11.88 |
| Energy contribution | -11.03 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1419737 101 - 27905053 --------CGUUGAGCCAGUGGCAGUUGCACAAGC-GUCCG---UUGCCCUCGAAGAAGCGAAAUCG----GGGGUUUAGGUCCAUUUCGGUGCUUCGCUGAGCAACAAUGCUGUGU --------......(((...)))....(((((.((-((..(---(((((..(((((.(.((((((.(----((........))))))))).).)))))..).))))).))))))))) ( -33.60, z-score = -0.31, R) >droVir3.scaffold_13047 11657266 98 + 19223366 ------UAAAAUGUGCCAGUUGC-UGGACGCCGGC-GCACA---UUGUCCUCGAGCCAGCGAAAUGC----G--AC-UAAAUGAAUUUCGGUGG-CAAGCGGACAAUAAUGCUGAGU ------......(((((((...)-))).)))((((-(...(---((((((.(..((((.((((((..----.--..-.......)))))).)))-)..).)))))))..)))))... ( -36.00, z-score = -2.47, R) >droGri2.scaffold_14830 2848172 107 + 6267026 UAUAACAACAAUGUGCCAGUUGCAAGUGCACAGAC-GCACU---UUGCCAGCGAGUCAGCGAAAUGA----U-UGCAUUCACACAUUUCGGUGG-CAAGCUGGCAAUAAUGCUGAUU ...........((((((........).)))))...-(((..---((((((((..((((.(((((((.----.-((....))..))))))).)))-)..))))))))...)))..... ( -38.30, z-score = -2.83, R) >droMoj3.scaffold_6540 30624340 100 + 34148556 ------CAACAUGUGCCAGUUGCAAGGACGCAGGC-GCACA---UUGUCUGCGAGCCAGCGAAAUGU----G-AAC-GAAAUGCAUUUCGGUGA-CGGGCAGACAAUAAUGCUGAGU ------((.((((((((...(((......))))))-))..(---((((((((..(.((.((((((((----.-...-.....)))))))).)).-)..))))))))).))).))... ( -37.50, z-score = -2.42, R) >droWil1.scaffold_181108 3221591 96 + 4707319 ---------CUUGUGCCAGUGGCAAUUGGGCAAAU-GUAUACGUUAAUCCUCGAAUCAGCGAAAU----------UUCCAUG-AAUUUCGGUGUUUCGCAGGAUAACAAUGCUGAGU ---------....((((...))))(((.(((((((-(....)))).(((((((((.((.((((((----------((....)-))))))).)).)))).))))).....)))).))) ( -30.00, z-score = -2.58, R) >droPer1.super_3 1229264 104 + 7375914 ---------CCUGUGCCAGUGGCUGCUGCGCAAUC-GUACG---UCACCUUCGGUUCAGCGAAAUCAGACCGAUAUCCGAUCCGAUUUCGGUGUUUCCGCAGGCAACAAUGCUGAGU ---------..(((((.(((....))))))))...-....(---(..(((.(((..((.(((((((.((.((.....)).)).))))))).))...))).)))..)).......... ( -32.10, z-score = -0.61, R) >dp4.chr2 7418864 104 + 30794189 ---------CCUGUGCCAGUGGCUGCUGCGCAAUC-GUACG---UCACCUUCGGUUCAGCGAAAUCAGACCGAUAUCCGAUACGAUUUCGGUGUUCCCGCAGGCAACAAUGCUGAGU ---------..((((((.((((..((..((.((((-(((((---......((((((...........))))))....)).))))))).))..))..)))).))).)))......... ( -30.20, z-score = -0.05, R) >droAna3.scaffold_13088 63453 103 + 569066 -------UGUAUGUGCCGGUGACAACUGCACAGGCUCCCCA---UUGUCCUCGAGGCAGCGAAAUAA----AAUACAUGCGUUCAUUUCGGUGAUCCCAAGGACAAUAAUGCUGAUG -------....(((((.(((....))))))))(((.....(---(((((((.(.((((.((((((..----...((....))..)))))).))..))).))))))))...))).... ( -28.30, z-score = -0.78, R) >droEre2.scaffold_4770 1689824 101 - 17746568 --------UGUUGAGCCAGUGGCAGCUGCACACGC-GUCCG---UUGCCAUCGAAGCAGCGAAAUCG----GUGGUUGAUGACCAUUUCGGUGUUUCGCUGGGCAACAAUGCUGAGU --------(((((..((((((((((((((....))-)...)---)))))...((((((.((((((.(----((........))))))))).))))))))))).)))))......... ( -41.40, z-score = -1.96, R) >droYak2.chr3R 17383645 101 - 28832112 --------UGUUGAGCCGGUGGCAGCUGCACAUGC-GUCCG---UUGCCCACGAAGCAGCGAAAUCG----GUGGUUUAGGUCCAUUUCGGUGUUUCGCUGGGCAACAAUGCUGUGU --------......(((...)))....(((((.((-((..(---((((((((((((((.((((((.(----(..........)))))))).))))))).)))))))).))))))))) ( -45.40, z-score = -3.01, R) >droSec1.super_6 1534622 101 - 4358794 --------CGUUGAGCCAGUGACAGUUGCACAAGC-GUCCG---UUGCCCUCGAAGCAGCGAAAUCG----GGGGUUUAUGUCCAUUUCGGUGCUUCGCUGGGCAACAAUGCUGUGU --------.(((........)))....(((((.((-((..(---((((((.(((((((.((((((.(----((.(....).))))))))).)))))))..))))))).))))))))) ( -45.10, z-score = -3.66, R) >droSim1.chr3R 1456175 101 - 27517382 --------CGUUGAGCCAGUGACAGUUGCACACGC-GUCCG---UUGCCCUCGAAGCAGCGAAAUCG----GGGGUUUAGGUCCAUUUCGGUGCUUCGCUGGGCAACAAUGCUGUGU --------.(((........)))....(((((.((-((..(---((((((.(((((((.((((((.(----((........))))))))).)))))))..))))))).))))))))) ( -44.70, z-score = -3.06, R) >consensus ________CGUUGUGCCAGUGGCAGCUGCACAAGC_GUACG___UUGCCCUCGAAGCAGCGAAAUCG____G_GGUUUAAGUCCAUUUCGGUGUUUCGCCGGGCAACAAUGCUGAGU ................(((((((.............))).......((((......((.((((((...................)))))).)).......))))......))))... (-11.88 = -11.03 + -0.85)
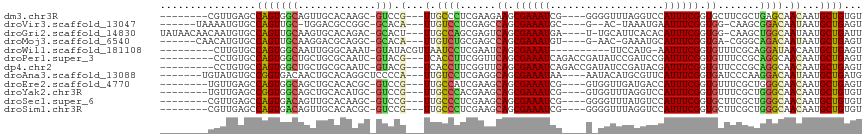
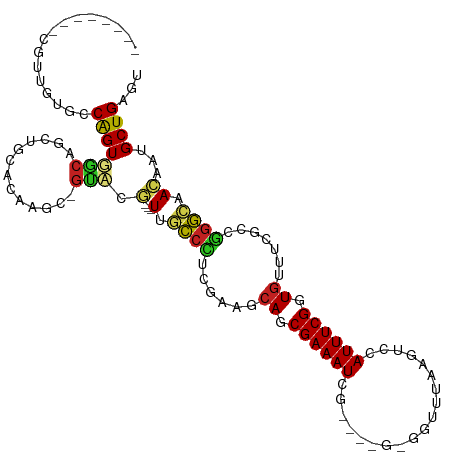
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:39 2011