| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,399,024 – 1,399,136 |
| Length | 112 |
| Max. P | 0.668613 |

| Location | 1,399,024 – 1,399,136 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.83 |
| Shannon entropy | 0.30439 |
| G+C content | 0.59665 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -29.86 |
| Energy contribution | -32.26 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1399024 112 - 27905053 GUUGGUGCCGAGUUGCGAUGUGGCGAAGUGACGUACUGCGCCCUCGUCACACACUCUUCCACAUUGGAUCGGAUGUGGCACCAGUGCGGUGGCU-ACAGAACGGCAUUGACGC ((..(((((((((((.((((.((((.((((...)))).))))..)))).)).)))).(((.....)))..(..(((((((((.....))).)))-)))...))))))..)).. ( -46.80, z-score = -1.76, R) >droSim1.chr3R 1434413 112 - 27517382 GUUGGUGCCGAGUGGCGAAGUGACGAGGUGACGUACUGCGCCCUCGUCACACACUCUCCCACAUCGGAUCGGAUGUGGCACCAGUGCGGUGGCU-ACAGAACGGCAUUGACGC ((..((((((.(((((...(((((((((.(.((.....))))))))))))........(((((((......)))))))((((.....)))))))-))....))))))..)).. ( -55.70, z-score = -3.65, R) >droSec1.super_6 1513749 112 - 4358794 GUUGGGGCCAAGUGGAGAAGUGACGAAGUGACGUACUGCGCCCUCGUCACACACUCUCCCACAUCGAAUCGGAUGUGGCACCAGUGCGGUGGCU-ACAGAACGGCAUUGACGC ((..(.(((..(((((((.(((((((.(((........)))..)))))))....))).))))........(..(((((((((.....))).)))-)))...)))).)..)).. ( -40.90, z-score = -0.71, R) >droYak2.chr3R 17362934 103 - 28832112 ----AUAGCGUAUUGC---GUGGCGAAGUGACGAACUACGUCCUCGUCACCCACACUCUCACAC---AUCGGAUGUGGCACCACUGCGGUGGCUCCCAGAACGGCAUUGACGC ----...((((..(((---(((((((.(.((((.....))))))))))))(((((.(((.....---...))))))))((((.....))))............)))...)))) ( -34.70, z-score = -0.69, R) >droEre2.scaffold_4770 1668732 99 - 17746568 GUUGGUGCUGAGUGG--------CGAAGUGACGUACUACGCCCUCGUCACACACUCUCCCACU-----UCGGAUGUGGCACCACUGCGGUGGCU-CCACAACGGCAUUGACGC ((..((((((.((((--------(((.(((........)))..)))))))......(((....-----..)))(((((..((((....))))..-))))).))))))..)).. ( -40.20, z-score = -1.89, R) >consensus GUUGGUGCCGAGUGGCGA_GUGACGAAGUGACGUACUGCGCCCUCGUCACACACUCUCCCACAU_G_AUCGGAUGUGGCACCAGUGCGGUGGCU_ACAGAACGGCAUUGACGC ((..((((((((((.....(((((((.(.(.((.....)))).))))))).)))))..(((((((......)))))))((((.....))))...........)))))..)).. (-29.86 = -32.26 + 2.40)

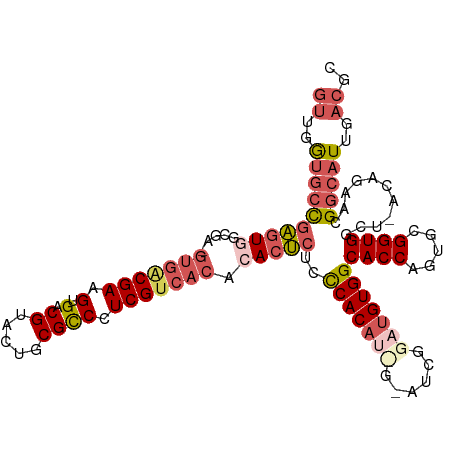
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:34 2011