| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,394,176 – 1,394,273 |
| Length | 97 |
| Max. P | 0.745583 |

| Location | 1,394,176 – 1,394,273 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.76304 |
| G+C content | 0.46002 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -10.67 |
| Energy contribution | -10.03 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1394176 97 + 27905053 GGGGCUUCGAGAUCUCCGCCUCGAAGCGUGAGAGCUCAUCCUCGAUGUUGCGACGCUUUGUACCCAUUUGAAGUGGGGUUUUCUCUUUUAAAAAAUC----------------- ...((((((((........))))))))(.((((((((....(((......)))(((((((........))))))))))))))).)............----------------- ( -29.20, z-score = -0.96, R) >droSim1.chr3R 1429676 97 + 27517382 GAGGCUUCGAGAUCUCCGCCUCGAAGCGUGAGAGCUCAUCCUCGAUGUUGCGACGCUUUGUACCCAUUUUAAAUGGGUUUGGCUCUUUUAAAAAAUC----------------- (((((((((((........))))))))((....)).....)))........((.(((....((((((.....))))))..)))))............----------------- ( -29.60, z-score = -1.82, R) >droYak2.chr3R 17357865 108 + 28832112 GAGGCUUCGAGAUCUCCGCCUCGAAGCGUGAGAGCUCAUCCUCGAUGUUCCGACGCUUUGUACCCAUUUAAAGUGGGUUUGGGGCUACUAGAAAACUUGAAGAUUCUC------ (((.(((((((.(((..((((((((((((..((((((......)).))))..)))))))..((((((.....))))))..)))))....)))...))))))).)))..------ ( -39.00, z-score = -2.54, R) >droEre2.scaffold_4770 1663827 108 + 17746568 GAGGCUUCGAGAUCUCCGCCUCGAAGCGAGAGAGCUCAUCCUCGAUGUUGCGACGCUUUGUACCCAUUUAACAUAGGUUUGGGGCUAUGAGAAAACUGGAAGAUUCUC------ (((.((((.((.((((.((((((((.((((.((.....)))))((((.(((((....)))))..)))).......).))))))))...))))...)).)))).)))..------ ( -30.70, z-score = -0.02, R) >droAna3.scaffold_13250 3507089 90 - 3535662 GAGGCUUUGAUAUCUCCGCCUCAAAGCGGGACAACUCGUCCUCGAUAUUUCGACGUUUUGUGCCCAUCGAU-GUUUUUUUUAAGGAUGCUC----------------------- (((((...(....)...)))))..((((((((((..((((...((....))))))..)))).)))......-(((((.....)))))))).----------------------- ( -22.80, z-score = -0.91, R) >dp4.chr2 9588925 105 - 30794189 GGGGCUUGGAGAUCUCAGCUUCGAAGCGAGACAGCUCGUCCUCGAUAUUGCGACGUUUCGUACCCAUUUAUUGAUAUCUUAAUGGAUAAAUAAUUUUU-AACUCUC-------- .(((..(((((.((.(((..((((.(((((....)))))..))))..))).))..)))))..)))(((((((.((......)).))))))).......-.......-------- ( -25.70, z-score = -0.95, R) >droPer1.super_3 3394483 106 - 7375914 GGGGCUUGGAGAUCUCAGCUUCGAAGCGAGACAGCUCGUCCUCGAUAUUGCGACGUUUCGUACCCAUUUAUUGAUAUCUUAAUGGAUAAAUAAUUUUUUAACUCUC-------- .(((..(((((.((.(((..((((.(((((....)))))..))))..))).))..)))))..)))(((((((.((......)).)))))))...............-------- ( -25.70, z-score = -0.91, R) >droWil1.scaffold_181130 13222044 84 + 16660200 GCGGCUUGGAAAUUUCUGCUUCAAAGCGCGACAAUUCGUCCUCUAUGUGACGACGUUUUGUACCCAUUUUUUAA-AUAUCAACUC----------------------------- (((.(((.(((........))).)))))).((((..((((.((.....)).))))..)))).............-..........----------------------------- ( -17.70, z-score = -1.57, R) >droVir3.scaffold_13047 16652971 112 + 19223366 GCGGCUUAGAGAUCUCCGCUUCGAAGCGCGAGAGCUCAUCUUCGAUAUUGCGACGUUUUGUGCCCAUUUCGAA-AGGCGUA-CGCUAUUGUUGAGAUGUCAACAAUAAAAAUUC ..(((...(((.((((((((....)))).)))).)))((((.(((((..((((((((((.............)-)))))).-)))...)))))))))))).............. ( -33.22, z-score = -1.16, R) >droMoj3.scaffold_6540 12960547 112 + 34148556 GCGGCUUUGAGAUCUCUGCUUCAAAGCGUGAGAGCUCAUCUUCGAUGUUACGACGUUUUGUCCCCAUUUCAGA-GGCUAAACUUCUAAUCAAACGAUUUCAAGUAAAAUAGUC- ..(.(((((((((........((((((((...(((((......)).)))...)))))))).....))))))))-).)...((((..((((....))))..)))).........- ( -24.12, z-score = -0.18, R) >droGri2.scaffold_15116 1618133 98 + 1808639 GCGGCUUGGAGAUCUCCGCCUCAAAUCGAGAGAGCUCGUCUUCGAUAUUGCGACGUUUUGUGCCCAUUUC-AA-AGGCGUAACGUUAUUUUCGUU-UGCCU------------- ..(((((((((((....(((((.......))).))..))))))))......((((((...((((......-..-.)))).)))))).........-.))).------------- ( -24.50, z-score = 0.23, R) >consensus GAGGCUUCGAGAUCUCCGCCUCGAAGCGAGAGAGCUCAUCCUCGAUGUUGCGACGUUUUGUACCCAUUUAAAAUGGGUUUAACGCUAUUAUAAAAUU___A_____________ ..(((...((....)).))).(((((((.((....))....(((......))))))))))...................................................... (-10.67 = -10.03 + -0.64)

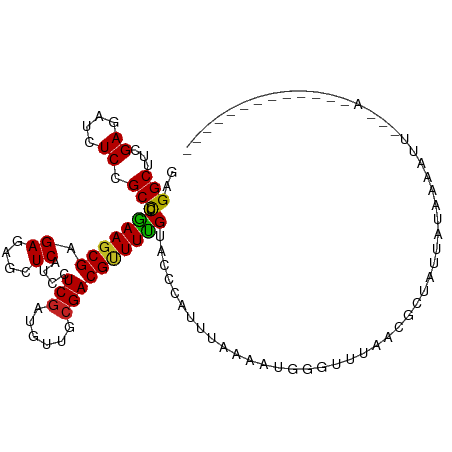

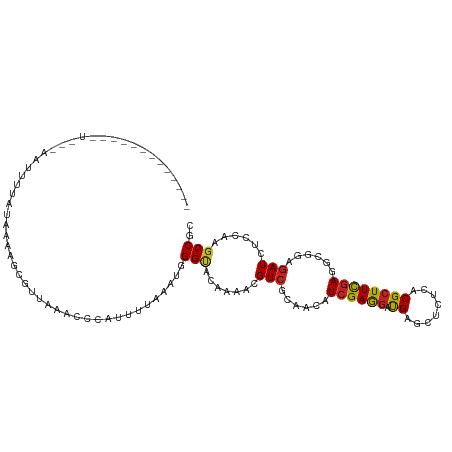
| Location | 1,394,176 – 1,394,273 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.76304 |
| G+C content | 0.46002 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -10.14 |
| Energy contribution | -9.66 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1394176 97 - 27905053 -----------------GAUUUUUUAAAAGAGAAAACCCCACUUCAAAUGGGUACAAAGCGUCGCAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCGAAGCCCC -----------------............((((....((((.......))))..((...(.(((......))).).))...))))..(((((((((......)))))))))... ( -23.70, z-score = -0.28, R) >droSim1.chr3R 1429676 97 - 27517382 -----------------GAUUUUUUAAAAGAGCCAAACCCAUUUAAAAUGGGUACAAAGCGUCGCAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCGAAGCCUC -----------------...........(((((((.((((((.....))))))......(.(((......))).).)).)))))...(((((((((......)))))))))... ( -29.90, z-score = -2.32, R) >droYak2.chr3R 17357865 108 - 28832112 ------GAGAAUCUUCAAGUUUUCUAGUAGCCCCAAACCCACUUUAAAUGGGUACAAAGCGUCGGAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCGAAGCCUC ------(((.((((((..((.((((....((.....(((((.......))))).....))...)))).)).))))))...)))....(((((((((......)))))))))... ( -31.10, z-score = -0.92, R) >droEre2.scaffold_4770 1663827 108 - 17746568 ------GAGAAUCUUCCAGUUUUCUCAUAGCCCCAAACCUAUGUUAAAUGGGUACAAAGCGUCGCAACAUCGAGGAUGAGCUCUCUCGCUUCGAGGCGGAGAUCUCGAAGCCUC ------(((...((((.((...((((...((.....((((((.....)))))).....))..(((....((((((.((((....)))))))))).))))))).)).)))).))) ( -32.60, z-score = -1.44, R) >droAna3.scaffold_13250 3507089 90 + 3535662 -----------------------GAGCAUCCUUAAAAAAAAC-AUCGAUGGGCACAAAACGUCGAAAUAUCGAGGACGAGUUGUCCCGCUUUGAGGCGGAGAUAUCAAAGCCUC -----------------------((((...............-.((((((.........))))))......(.((((.....))))))))).(((((.((....))...))))) ( -23.90, z-score = -1.56, R) >dp4.chr2 9588925 105 + 30794189 --------GAGAGUU-AAAAAUUAUUUAUCCAUUAAGAUAUCAAUAAAUGGGUACGAAACGUCGCAAUAUCGAGGACGAGCUGUCUCGCUUCGAAGCUGAGAUCUCCAAGCCCC --------((((...-..........((((((((............)))))))).......((.((.(.((((((.((((....)))))))))).).)).))))))........ ( -22.10, z-score = -0.51, R) >droPer1.super_3 3394483 106 + 7375914 --------GAGAGUUAAAAAAUUAUUUAUCCAUUAAGAUAUCAAUAAAUGGGUACGAAACGUCGCAAUAUCGAGGACGAGCUGUCUCGCUUCGAAGCUGAGAUCUCCAAGCCCC --------((((..............((((((((............)))))))).......((.((.(.((((((.((((....)))))))))).).)).))))))........ ( -22.10, z-score = -0.51, R) >droWil1.scaffold_181130 13222044 84 - 16660200 -----------------------------GAGUUGAUAU-UUAAAAAAUGGGUACAAAACGUCGUCACAUAGAGGACGAAUUGUCGCGCUUUGAAGCAGAAAUUUCCAAGCCGC -----------------------------..........-.........(.(.((((..((((.((.....)).))))..))))).)((((.((((......)))).))))... ( -16.40, z-score = -0.19, R) >droVir3.scaffold_13047 16652971 112 - 19223366 GAAUUUUUAUUGUUGACAUCUCAACAAUAGCG-UACGCCU-UUCGAAAUGGGCACAAAACGUCGCAAUAUCGAAGAUGAGCUCUCGCGCUUCGAAGCGGAGAUCUCUAAGCCGC .......(((((((((....)))))))))(((-...((.(-(((((.((..((((.....)).)).)).))))))..(((.((((.((((....)))))))).)))...))))) ( -31.40, z-score = -1.23, R) >droMoj3.scaffold_6540 12960547 112 - 34148556 -GACUAUUUUACUUGAAAUCGUUUGAUUAGAAGUUUAGCC-UCUGAAAUGGGGACAAAACGUCGUAACAUCGAAGAUGAGCUCUCACGCUUUGAAGCAGAGAUCUCAAAGCCGC -(((..(((((.(..((....))..).)))))((((.(((-((......)))).).))))))).............((((.((((..(((....))).)))).))))....... ( -23.60, z-score = 0.41, R) >droGri2.scaffold_15116 1618133 98 - 1808639 -------------AGGCA-AACGAAAAUAACGUUACGCCU-UU-GAAAUGGGCACAAAACGUCGCAAUAUCGAAGACGAGCUCUCUCGAUUUGAGGCGGAGAUCUCCAAGCCGC -------------.(((.-((((.......))))..((((-..-.....)))).......(((.(((.((((((((.....))).)))))))).)))(((....)))..))).. ( -26.00, z-score = -0.77, R) >consensus _____________U___AAUUUUAUAAAAGCGUUAAACCCAUUUUAAAUGGGUACAAAACGUCGCAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCCAAGCCGC ..................................................(((.......(((......((((((.((........))))))))......)))......))).. (-10.14 = -9.66 + -0.49)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:33 2011