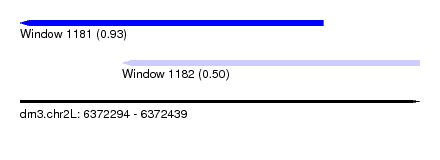
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,372,294 – 6,372,439 |
| Length | 145 |
| Max. P | 0.931034 |

| Location | 6,372,294 – 6,372,404 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.11 |
| Shannon entropy | 0.25066 |
| G+C content | 0.40472 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.41 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
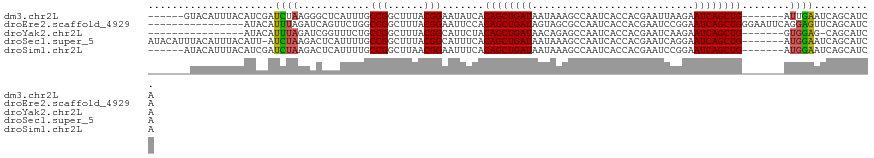
>dm3.chr2L 6372294 110 - 23011544 CUUUACGGAAUAUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUUAAGAAUCAGCUG-------AUUGAAUCAGCAUCAAAGCUCAGUGCUUUUUAAACCGUAAAC---UAAUCUAGAC .(((((((.......((((((((...........................))))))))-------.(((((..(((((.........))))).))))).))))))).---.......... ( -22.23, z-score = -2.50, R) >droEre2.scaffold_4929 15298215 120 - 26641161 CUUUACGGAAUUCCACAGCUGAUAGUAGCGCCAAUCACCACGAAUCCGGAAUCAGCUGGGAAUUCAGGAGUUCAGCAUCAAAGCUCAGUGCUUCUUAAACCGUAAACACAUAAUCUUGAC .(((((((((((((.((((((((.(.....)......((........)).)))))))))))))))((((((..(((......)))....)))))).....)))))).............. ( -33.90, z-score = -3.21, R) >droYak2.chr2L 15790173 109 + 22324452 CUUUACGGCAUUCUACAGCUGAUAACAGAGCCAAUCACCACGAAUCAAGAAUCAGCUG--------GUGGAGCAGCAUCAAAGCUCAGUGCUUUUUAAACCGUAAUC---CUAACUUAAC ..((((((..(((((((((((((....((..(.........)..))....))))))).--------)))))).......(((((.....))))).....))))))..---.......... ( -24.30, z-score = -1.52, R) >droSec1.super_5 4440223 113 - 5866729 CUUUACGGCAUUUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUCAGGAAUCAGCUG-------AUGGAAUCAGCAUCAAAGCUCAGUGCUUUUUAAACCGUAAACUAAUAAUCUAGAC .(((((((.((((((((((((((......(.....).((........)).))))))))-------.)))))).......(((((.....))))).....)))))))(((......))).. ( -25.10, z-score = -2.46, R) >droSim1.chr2L 6176448 113 - 22036055 CUUAACGGAAUUUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUCCGGAAUCAGCUG-------AUGGAAUCAGCAUCAAAGCUCAGUGCUUUUUAAACCGUAAACUAAUAAUCUAGAC ....((((.((((((((((((((......(.....).((........)).))))))))-------.)))))).......(((((.....))))).....))))...(((......))).. ( -23.10, z-score = -1.95, R) >consensus CUUUACGGAAUUUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUCAGGAAUCAGCUG_______AUGGAAUCAGCAUCAAAGCUCAGUGCUUUUUAAACCGUAAAC__AUAAUCUAGAC .(((((((.......((((((((...........................)))))))).....................(((((.....))))).....))))))).............. (-18.81 = -19.41 + 0.60)

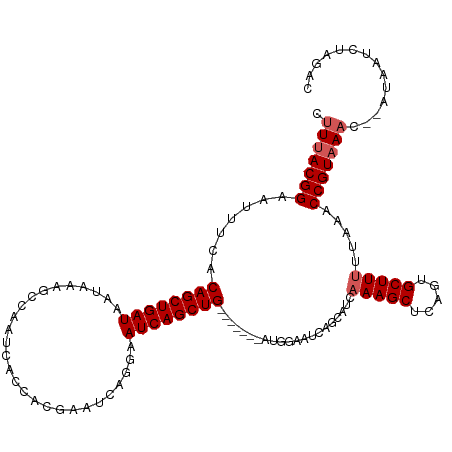
| Location | 6,372,331 – 6,372,439 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 74.06 |
| Shannon entropy | 0.42736 |
| G+C content | 0.42524 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -12.81 |
| Energy contribution | -12.65 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 6372331 108 - 23011544 ------GUACAUUUACAUCGAUCUAAGGGCUCAUUUGCCGGCUUUACGGAAUAUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUUAAGAAUCAGCUG-------AUUGAAUCAGCAUCA ------...........(((.......(((......)))(((((((.....(((((....))))).)))))))........)))...........((((-------((...)))))).... ( -24.20, z-score = -1.97, R) >droEre2.scaffold_4929 15298255 105 - 26641161 ----------------AUACAUUUAGAUCAGUUCUGGCCGGCUUUACGGAAUUCCACAGCUGAUAGUAGCGCCAAUCACCACGAAUCCGGAAUCAGCUGGGAAUUCAGGAGUUCAGCAUCA ----------------.......((((.....))))((.((((((...(((((((.((((((((.(.....)......((........)).))))))))))))))).))))))..)).... ( -28.30, z-score = -1.47, R) >droYak2.chr2L 15790210 97 + 22324452 ----------------AUACAUUUAGAUCGGUUUCUGCCGGCUUUACGGCAUUCUACAGCUGAUAACAGAGCCAAUCACCACGAAUCAAGAAUCAGCUG-------GUGGAG-CAGCAUCA ----------------.........((((.((((((((((......))))).....((((((((....((..(.........)..))....))))))))-------..))))-).).))). ( -25.50, z-score = -1.54, R) >droSec1.super_5 4440263 113 - 5866729 AUACAUUUACAUUUACAUU-AUCUAAGACUCAUUUUGCCGGCUUUACGGCAUUUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUCAGGAAUCAGCUG-------AUGGAAUCAGCAUCA ...................-...............(((((......)))))(((((((((((((......(.....).((........)).))))))))-------.)))))......... ( -21.90, z-score = -1.89, R) >droSim1.chr2L 6176488 108 - 22036055 ------AUACAUUUACAUCGAUCUAAGACUCAUUUUGCCGGCUUAACGGAAUUUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUCCGGAAUCAGCUG-------AUGGAAUCAGCAUCA ------.............(((((.............(((......))).((((((((((((((......(.....).((........)).))))))))-------.)))))).)).))). ( -20.10, z-score = -0.70, R) >consensus _______UACAUUUACAUACAUCUAAGACUCAUUUUGCCGGCUUUACGGAAUUUCACAGCUGAUAAUAAAGCCAAUCACCACGAAUCAGGAAUCAGCUG_______AUGGAAUCAGCAUCA .....................((((............(((......))).......((((((((...........................))))))))........)))).......... (-12.81 = -12.65 + -0.16)
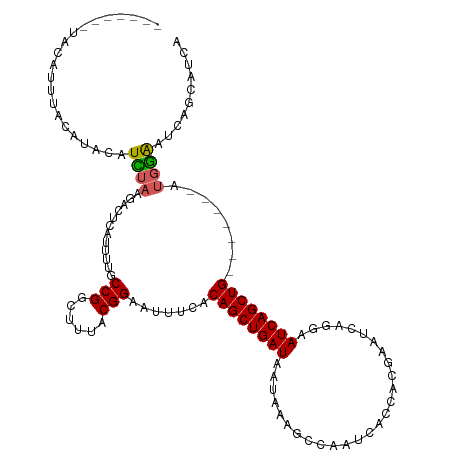
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:08 2011