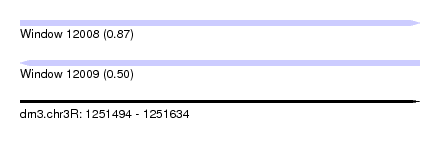
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,251,494 – 1,251,634 |
| Length | 140 |
| Max. P | 0.874365 |

| Location | 1,251,494 – 1,251,634 |
|---|---|
| Length | 140 |
| Sequences | 7 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 61.38 |
| Shannon entropy | 0.75659 |
| G+C content | 0.60105 |
| Mean single sequence MFE | -49.79 |
| Consensus MFE | -17.19 |
| Energy contribution | -19.61 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
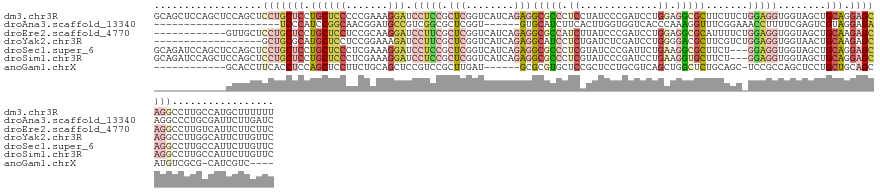
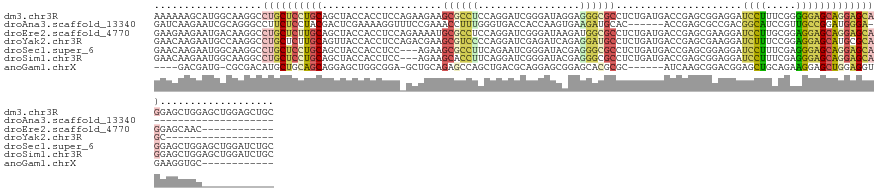
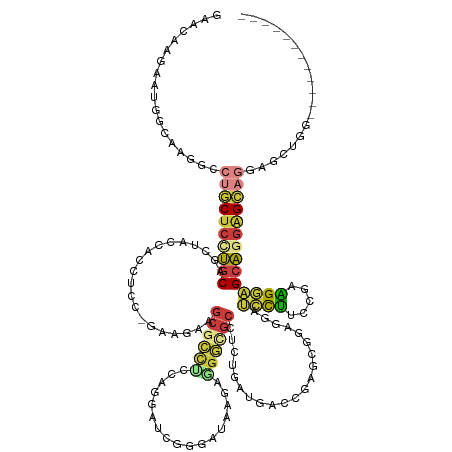
>dm3.chr3R 1251494 140 + 27905053 GCAGCUCCAGCUCCAGCUCCUGCUCCUGCUCCCCCGAAAGGAUCCUCCGCUCGGUCAUCAGAGGCGCCCUCCUAUCCCGAUCCUGGAGGCGCUUCUUCUGGAGGUGGUAGCUGCAGGAGCAGGCCUUGCCAUGCUUUUUU (((((((((.((((((.....((....))....((((..((.....))..)))).....(((((((((.(((............)))))))))))).)))))).))).))))))((((((((((...))).))))))).. ( -64.30, z-score = -2.85, R) >droAna3.scaffold_13340 19760435 113 - 23697760 ---------------------UCCCAUCCGGCAACGGAUGCCGUCGGCGCUCGGU------GUGCAUCUUCACUUGGUGGUCACCCAAAGGUUUCGGAAACCUUUUCGAGUCGUAGGAGAAGGCCCUGCGAUUCUUGAUC ---------------------...((((((....)))))).......((((.(((------(........)))).))))((((...(((((((.....)))))))..((((((((((.......)))))))))).)))). ( -41.00, z-score = -1.27, R) >droEre2.scaffold_4770 1521334 128 + 17746568 ------------GUUGCUCCUGCUCCUGCUCCUCCGCAAGGAUCCUUCGCUCGGUCAUCAGAGGCGCCAUCUUAUCCCGAUCCUGGAGGCGCAUUUUCUGGAGGUGGUAGCUGCAAGAGCAGGCCUUGUCAUUCUUCUUC ------------.((((((.(((..(((((((((((((((....))).)).........(((.(((((.(((............))))))))....)))))))).)))))..))).)))))).................. ( -43.80, z-score = -0.45, R) >droYak2.chr3R 1649736 122 + 28832112 ------------------GCUGCGCAUGCUCCUCCGGAAAGAUCCUUCGCUCGGUCAUCAGAGGCAUCCUCUGAUCUCGAUCCUGGGGACGCUUCGUCUGGAGGUGGUAACUGCAAGAGCAGGCCUUGGCAUUCUUGUUC ------------------(((..((.((((((((((((..((((........))))....(((((.(((((.(((....)))..))))).))))).)))))))).)))).((((....))))))...))).......... ( -45.70, z-score = -0.88, R) >droSec1.super_6 1370530 137 + 4358794 GCAGAUCCAGCUCCAGCUCCUGCUCCUGCUCCCUCGAAAGGAUCCUCCGCUCGGUCAUCAGAGGCGCCCUCGUAUCCCGAUUCUGAAGGCGCUUCU---GGAGGUGGUAGCUGCAGGAGCAGGCCUUGCCAUUCUUGUUC ((((...........((((((((..((((..((((((..((.....))..))......((((((((((.(((...........))).)))))))))---)))))..))))..)))))))).(((...)))....)))).. ( -58.90, z-score = -2.37, R) >droSim1.chr3R 1250392 137 + 27517382 GCAGAUCCAGCUCCAGCUCCUGCUCCUGCUCCCUCGAAAGGAUCCUCCGCUCGGUCAUCAGAGGCGCCCUCGUAUCCCGAUCCUGAAGGUGCUUCU---GGAGGUGGUAGCUGCAGGAGCAGGCCUUGCCAUUCUUGUUC ((((...........((((((((..((((..((((((..((.....))..))......((((((((((.(((...........))).)))))))))---)))))..))))..)))))))).(((...)))....)))).. ( -57.00, z-score = -1.99, R) >anoGam1.chrX 14832065 116 - 22145176 ------------GCACCUUCACCUCCAGCUCCUUCUGCAGCUCCGUCCGCUUGAU------GCGCGUGCUCCGCUCCUGCGUCAGCUGGCUCUGCAGC-UCCGCCAGCUCCUGCUGCAGCAUGUCGCG-CAUCGUC---- ------------..............((((.(....).))))..........(((------(((((.((...(((.(.(((..(((((((........-...)))))))..))).).)))..))))))-))))...---- ( -37.80, z-score = -0.33, R) >consensus ____________CCAGCUCCUGCUCCUGCUCCCCCGAAAGGAUCCUCCGCUCGGUCAUCAGAGGCGCCCUCCUAUCCCGAUCCUGGAGGCGCUUCUUC_GGAGGUGGUAGCUGCAGGAGCAGGCCUUGCCAUUCUUGUUC ..................(((((((.((((((.......))).(((((.(((........)))(((((.((.............)).))))).......)))))........))).)))))))................. (-17.19 = -19.61 + 2.42)
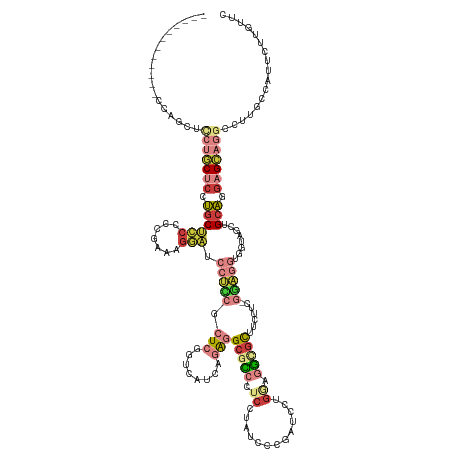
| Location | 1,251,494 – 1,251,634 |
|---|---|
| Length | 140 |
| Sequences | 7 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 61.38 |
| Shannon entropy | 0.75659 |
| G+C content | 0.60105 |
| Mean single sequence MFE | -47.39 |
| Consensus MFE | -20.54 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.70 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 1251494 140 - 27905053 AAAAAAGCAUGGCAAGGCCUGCUCCUGCAGCUACCACCUCCAGAAGAAGCGCCUCCAGGAUCGGGAUAGGAGGGCGCCUCUGAUGACCGAGCGGAGGAUCCUUUCGGGGGAGCAGGAGCAGGAGCUGGAGCUGGAGCUGC .....(((.((((.((.(((((((((((.(....).(((((..(((.....(((((....((((....(((((...))))).....))))..)))))...)))..))))).)))))))))))..))...))))..))).. ( -61.40, z-score = -0.94, R) >droAna3.scaffold_13340 19760435 113 + 23697760 GAUCAAGAAUCGCAGGGCCUUCUCCUACGACUCGAAAAGGUUUCCGAAACCUUUGGGUGACCACCAAGUGAAGAUGCAC------ACCGAGCGCCGACGGCAUCCGUUGCCGGAUGGGA--------------------- ......((.(((.((((.....)))).))).))..(((((((.....)))))))((....)).(((.(((......)))------.(((.((...(((((...)))))))))).)))..--------------------- ( -33.40, z-score = 0.36, R) >droEre2.scaffold_4770 1521334 128 - 17746568 GAAGAAGAAUGACAAGGCCUGCUCUUGCAGCUACCACCUCCAGAAAAUGCGCCUCCAGGAUCGGGAUAAGAUGGCGCCUCUGAUGACCGAGCGAAGGAUCCUUGCGGAGGAGCAGGAGCAGGAGCAAC------------ .................(((((((((((.((.................)).(((((....((((....(((.(....)))).....))))((.(((....)))))))))).)))))))))))......------------ ( -43.53, z-score = -0.87, R) >droYak2.chr3R 1649736 122 - 28832112 GAACAAGAAUGCCAAGGCCUGCUCUUGCAGUUACCACCUCCAGACGAAGCGUCCCCAGGAUCGAGAUCAGAGGAUGCCUCUGAUGACCGAGCGAAGGAUCUUUCCGGAGGAGCAUGCGCAGC------------------ ......(.((((...((.((((....))))...)).(((((.((((...))))....(((..((.((((((((...))))))))..((.......)).))..)))))))).)))).).....------------------ ( -39.30, z-score = -0.63, R) >droSec1.super_6 1370530 137 - 4358794 GAACAAGAAUGGCAAGGCCUGCUCCUGCAGCUACCACCUCC---AGAAGCGCCUUCAGAAUCGGGAUACGAGGGCGCCUCUGAUGACCGAGCGGAGGAUCCUUUCGAGGGAGCAGGAGCAGGAGCUGGAGCUGGAUCUGC .....(((.((((.((.(((((((((((........(((((---....((((((((.............))))))))(((........))).))))).((((.....)))))))))))))))..))...))))..))).. ( -58.12, z-score = -1.61, R) >droSim1.chr3R 1250392 137 - 27517382 GAACAAGAAUGGCAAGGCCUGCUCCUGCAGCUACCACCUCC---AGAAGCACCUUCAGGAUCGGGAUACGAGGGCGCCUCUGAUGACCGAGCGGAGGAUCCUUUCGAGGGAGCAGGAGCAGGAGCUGGAGCUGGAUCUGC .....(((.((((.((.(((((((((((........(((((---.((((...))))....((((.....((((...))))......))))..))))).((((.....)))))))))))))))..))...))))..))).. ( -54.20, z-score = -0.68, R) >anoGam1.chrX 14832065 116 + 22145176 ----GACGAUG-CGCGACAUGCUGCAGCAGGAGCUGGCGGA-GCUGCAGAGCCAGCUGACGCAGGAGCGGAGCACGCGC------AUCAAGCGGACGGAGCUGCAGAAGGAGCUGGAGGUGAAGGUGC------------ ----...((((-((((...((((.(.((...(((((((...-........)))))))...)).).)))).....)))))------)))..(((..(...(((.(((......)))..)))...).)))------------ ( -41.80, z-score = 0.80, R) >consensus GAACAAGAAUGGCAAGGCCUGCUCCUGCAGCUACCACCUCC_GAAGAAGCGCCUCCAGGAUCGGGAUAAGAGGGCGCCUCUGAUGACCGAGCGGAGGAUCCUUCCGAAGGAGCAGGAGCAGGAGCUGG____________ ..................((((((((((....................((((((.................)))))).....................((((.....))))))))))))))................... (-20.54 = -21.14 + 0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:24 2011