
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,213,940 – 1,214,021 |
| Length | 81 |
| Max. P | 0.998355 |

| Location | 1,213,940 – 1,214,021 |
|---|---|
| Length | 81 |
| Sequences | 13 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 95.57 |
| Shannon entropy | 0.10168 |
| G+C content | 0.54606 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -29.87 |
| Energy contribution | -29.73 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.96 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
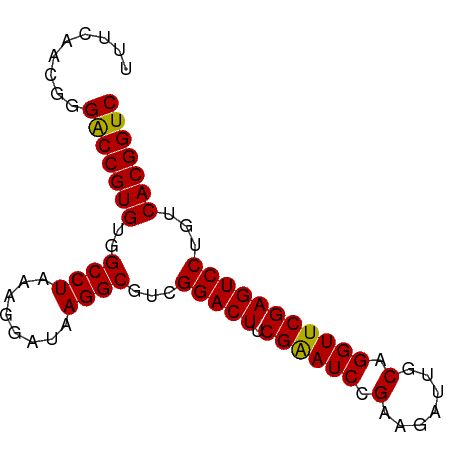
>dm3.chr3R 1213940 81 + 27905053 UUUCAACCGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -1.95, R) >droSim1.chr3R 1232427 81 + 27517382 UUUCAACCGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -1.95, R) >droSec1.super_6 1331373 81 + 4358794 UUUCAACCGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -1.95, R) >droYak2.chr3R 1603298 81 + 28832112 UGUCAGCCGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -1.67, R) >droEre2.scaffold_4770 1482974 81 + 17746568 UUUCAACGGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -1.89, R) >droAna3.scaffold_13340 19721832 81 - 23697760 UUUCAACAGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -2.34, R) >dp4.chr2 9229887 81 - 30794189 UUACAACGGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -2.00, R) >droPer1.super_3 3027637 81 - 7375914 UUACAACGGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -2.00, R) >droWil1.scaffold_181089 5049277 81 + 12369635 AUGCAAAAGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -2.52, R) >droVir3.scaffold_12855 9965840 81 + 10161210 UUACAACGGGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -1.90, R) >droGri2.scaffold_15074 3503702 81 - 7742996 UUACAACAGGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -29.80, z-score = -2.31, R) >droMoj3.scaffold_6540 11226855 81 - 34148556 UCGUAACAAGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) ( -30.00, z-score = -2.44, R) >anoGam1.chr2L 33896025 81 - 48795086 UGUCAGCGGGGCCGUGUGGCCUAAUGGAGAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))) ( -29.10, z-score = -0.56, R) >consensus UUUCAACGGGACCGUGUGGCCUAAAGGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUC .........(((((((..((((........))))...(((((.((((((.(.......).)))))))))))...))))))) (-29.87 = -29.73 + -0.14)

| Location | 1,213,940 – 1,214,021 |
|---|---|
| Length | 81 |
| Sequences | 13 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 95.57 |
| Shannon entropy | 0.10168 |
| G+C content | 0.54606 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -29.97 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
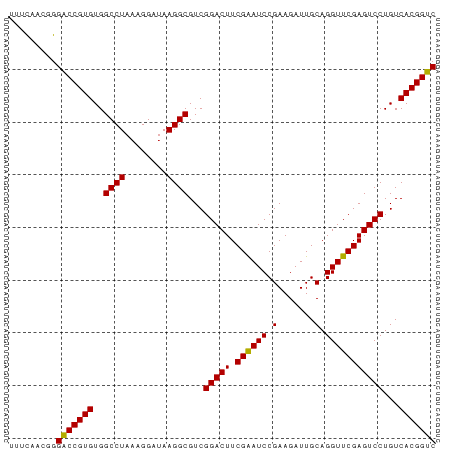
>dm3.chr3R 1213940 81 - 27905053 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCGGUUGAAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.16, R) >droSim1.chr3R 1232427 81 - 27517382 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCGGUUGAAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.16, R) >droSec1.super_6 1331373 81 - 4358794 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCGGUUGAAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.16, R) >droYak2.chr3R 1603298 81 - 28832112 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCGGCUGACA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.26, R) >droEre2.scaffold_4770 1482974 81 - 17746568 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCCGUUGAAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.68, R) >droAna3.scaffold_13340 19721832 81 + 23697760 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCUGUUGAAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.81, R) >dp4.chr2 9229887 81 + 30794189 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCCGUUGUAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.69, R) >droPer1.super_3 3027637 81 + 7375914 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCCGUUGUAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.69, R) >droWil1.scaffold_181089 5049277 81 - 12369635 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCUUUUGCAU (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -4.43, R) >droVir3.scaffold_12855 9965840 81 - 10161210 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCCCGUUGUAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.71, R) >droGri2.scaffold_15074 3503702 81 + 7742996 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCCUGUUGUAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -30.70, z-score = -3.77, R) >droMoj3.scaffold_6540 11226855 81 + 34148556 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCUUGUUACGA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... ( -31.10, z-score = -3.97, R) >anoGam1.chr2L 33896025 81 + 48795086 GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUCUCCAUUAGGCCACACGGCCCCGCUGACA ..(((((.((((.......))))......((((((.....)))))).((((........))))..)))))........... ( -22.90, z-score = -1.13, R) >consensus GACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCCGUUGAAA (((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......... (-29.97 = -30.12 + 0.15)
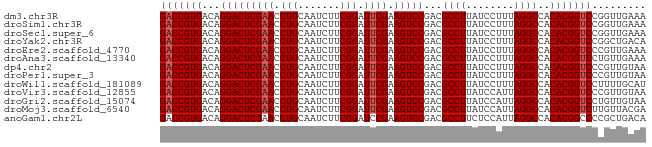


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:20 2011