| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,177,889 – 1,177,997 |
| Length | 108 |
| Max. P | 0.930936 |

| Location | 1,177,889 – 1,177,997 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Shannon entropy | 0.33078 |
| G+C content | 0.34620 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.47 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
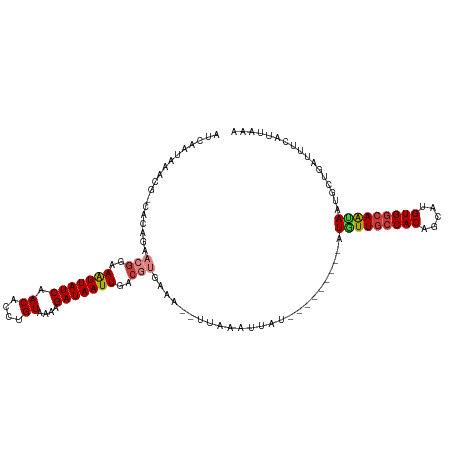
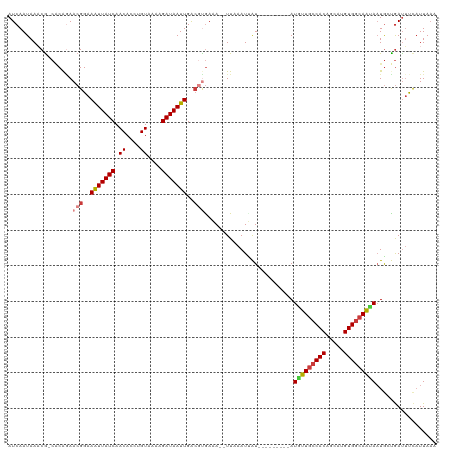
>dm3.chr3R 1177889 108 + 27905053 AUCAAUAAACG-CGCAGAACGGAAGUUAUCAACACCUGUAAGGGAUAAUUGACGUGAUA--UUAAAUGGU---------AUGUUGCCACAGCAGGUGGCAGCAUUGCCGAUUUCAUUAAA (((......((-((((((((....))).......(((....)))....))).))))...--......(((---------((((((((((.....))))))))).)))))))......... ( -32.10, z-score = -2.81, R) >droSec1.super_6 1296954 108 + 4358794 AUCAAUAAACG-CGCAGAACGGAAGUUAUCAACACCUGUAAGGGAUAAUUGACGUGAUA--UUAAAUGGU---------AUGUUGCCACAGCAUGUGGCAGCAAUGCCGAUUUCAUUAAA (((......((-((((((((....))).......(((....)))....))).))))...--......(((---------((((((((((.....)))))))).))))))))......... ( -32.50, z-score = -2.94, R) >droEre2.scaffold_4770 1447062 108 + 17746568 AUCAAUAAUCG-CACGGAACGGAAAUUAUCAACACAUGUAAGAGAUAAUUGACAUGAAA--UUAAAUCGU---------AUGUUGCCACAGCAGGUGGCAGGAAUGCCAAUAUCAUUAAA .(((....((.-....))..(..(((((((.............)))))))..).)))..--.......((---------((.(((((((.....)))))))..))))............. ( -18.32, z-score = 0.08, R) >droAna3.scaffold_13340 23354140 108 + 23697760 AUCAAUAAACG-CCCACAACGGAAAUUAUCAACACCUGUAAAAGAUAAUUGACGUGAAA--UUAAAUUAU---------AUGUUGCCACAGCGGGUGGCAAUAAUGUUGACUUUAUUUAA .((((((....-......(((..(((((((.............)))))))..)))....--.........---------.(((((((((.....))))))))).)))))).......... ( -25.32, z-score = -2.48, R) >dp4.chr2 9191343 108 - 30794189 AUCAAUAAACG-CACAGAACGGAAAUUAUCAACACCUGUAAAAGAUAAUUGACGCGAAA--UUAAAUUAU---------AUGUUGCCACAGCAUGUGGCAAUAAUGAUGAUUCCAUUAAA ...........-........((((.((((((.............((((((.........--...))))))---------.(((((((((.....))))))))).))))))))))...... ( -23.60, z-score = -2.68, R) >droPer1.super_3 2988050 108 - 7375914 AUCAAUAAACG-CACAGAACGGAAAUUAUCAACACCUGUAAAAGAUAAUUGACGCAAAA--UUAAAUUAU---------AUGUUGCCACAGCAUGUGGCAAUAAUGAUGAUUCCAUUAAA ...........-........((((.((((((.............((((((.........--...))))))---------.(((((((((.....))))))))).))))))))))...... ( -23.60, z-score = -2.83, R) >droWil1.scaffold_181130 3917732 116 - 16660200 AUCAAUAAUCAACACAAAACGGAAAUUAUCAACACCUGUAAACGAUAAUUGGGCUGAAAAAUUCAAUUAUUUAUUCAACAUAUUGCCACAACAUGUGAGAAUAAUA-UGAUUCCUUU--- ....................((((...................((((((((((.(....).)))))))))).......(((((((((((.....))).)..)))))-)).))))...--- ( -16.10, z-score = -0.83, R) >consensus AUCAAUAAACG_CACAGAACGGAAAUUAUCAACACCUGUAAAAGAUAAUUGACGUGAAA__UUAAAUUAU_________AUGUUGCCACAGCAUGUGGCAAUAAUGCUGAUUUCAUUAAA ..................(((..(((((((.((....))....)))))))..))).........................(((((((((.....)))))))))................. (-17.04 = -17.47 + 0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:16 2011