
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,351,659 – 6,351,769 |
| Length | 110 |
| Max. P | 0.999885 |

| Location | 6,351,659 – 6,351,769 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 59.38 |
| Shannon entropy | 0.73795 |
| G+C content | 0.53227 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
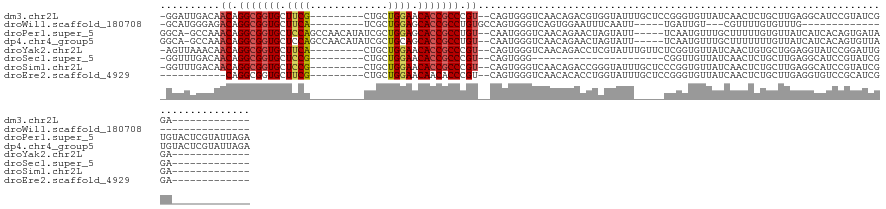
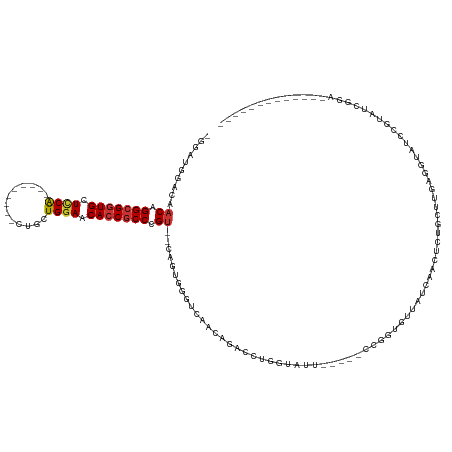
>dm3.chr2L 6351659 110 + 23011544 -GGAUUGACAACAGGCGGUGCUUCG---------CUGCUGGAACACCGCCCGU--CAGUGGGUCAACAGACGUGGUAUUUGCUCCGGGUGUUAUCAACUCUGCUUGAGGCAUCCGUAUCGGA------------- -.((((.((.((.(((((((.((((---------....)))).))))))).))--..)).))))....((..(((....(((..(((((............)))))..))).)))..))...------------- ( -35.80, z-score = 0.29, R) >droWil1.scaffold_180708 5398044 89 - 12563649 -GCAUGGGAGACAGGCGGUGCUUCA---------UCGCUGGAGCACCGCCUGUGCCAGUGGGUCAGUGGAAUUUCAAUU-----UGAUUGU---CGUUUUGUGUUUG---------------------------- -.(((.((..(((((((((((((((---------....))))))))))))))).)).))).(.((((.((((....)))-----).)))).---)............---------------------------- ( -38.70, z-score = -3.76, R) >droPer1.super_5 367517 127 - 6813705 GGCA-GCCAAACAGGCGGUGCUCCAGCCAACAUAUCGCUGGAGCACCGCCUGU--CAAUGGGUCAACAGAACUAGUAUU-----UCAAUGUUUGCUUUUUGUGUUAUCAUCACAGUGAUAUGUACUCGUAUUAGA (((.-.(((.(((((((((((((((((.........)))))))))))))))))--...))))))((((.((..((((..-----........))))..)).))))..........(((((((....))))))).. ( -50.10, z-score = -5.14, R) >dp4.chr4_group5 371564 127 - 2436548 GGCA-GCCAAACAGGCGGUGCUCCAGCCAACAUAUCGCUGCAGCACCGCCUGU--CAAUGGGUCAACAGAACUAGUAUU-----UCAAUGUUUGCUUUUUUUGUUAUCAUCACAGUGUUAUGUACUCGUAUUAGA (((.-.(((.((((((((((((.((((.........)))).))))))))))))--...))))))(((((((..((((..-----........))))..)))))))......((((((.....)))).))...... ( -42.00, z-score = -3.54, R) >droYak2.chr2L 15771633 110 - 22324452 -AGUUAAACAACAGGCGGUGCUUCA---------CUGCUGGAACACCGCCCGU--CAGUGGGUCAACAGACCUCGUAUUUGUUCUCGGUGUUAUCAACUGUGCUGGAGGUAUCCGGAUUGGA------------- -.........((((((((((...))---------))))((((((((((.....--(((.(((((....))))).....)))....))))))).))).)))).(((((....)))))......------------- ( -33.10, z-score = 0.12, R) >droSec1.super_5 4422289 88 + 5866729 -GGUUUGACAACAGGCGGUGCUCCG---------CUGCUGGAACACCGCCCGU--CAGUGGG----------------------CGGUUGUUAUCAACUCUGCUUGAGGCAUCCGUAUCGGA------------- -....(((((((.(((((((.((((---------....)))).)))))))(((--(....))----------------------)))))))))....(((.....)))...((((...))))------------- ( -34.10, z-score = -1.17, R) >droSim1.chr2L 6158513 110 + 22036055 -GGUUUGACAACAGGCGGUGCUCCG---------CUGCUGGAACACCGCCCGU--CAGUGGGUCAACAGACCGGGUAUUUGCUCCCGGUGUUAUCAACUCUGCUUGAGGCAUCCGUAUCGGA------------- -...(((((....(((((((.((((---------....)))).))))))).))--))).((((....(.((((((........)))))).).....))))(((.....)))((((...))))------------- ( -39.20, z-score = -0.39, R) >droEre2.scaffold_4929 15280183 100 + 26641161 -----------CAGGCGGUGCUUCG---------CUGCUGGAACAACACCCGU--CAGUGGGUCAACACACCUGGUAUUUGCUCCGGGUGUUAUCAACUCUGCUUGAGGUGUCCGCAUCGGA------------- -----------....((((((..((---------(((.(((........))).--)))))((.((..((((((((........))))))))......(((.....))).)).))))))))..------------- ( -37.00, z-score = -1.18, R) >consensus _GGAUGGACAACAGGCGGUGCUCCA_________CUGCUGGAACACCGCCCGU__CAGUGGGUCAACAGACCUGGUAUU_____CCGGUGUUAUCAACUCUGCUUGAGGUAUCCGUAUCGGA_____________ ..........((.(((((((.((((.............)))).))))))).)).................................................................................. (-14.32 = -14.38 + 0.06)
| Location | 6,351,659 – 6,351,769 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 135 |
| Reading direction | reverse |
| Mean pairwise identity | 59.38 |
| Shannon entropy | 0.73795 |
| G+C content | 0.53227 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -19.29 |
| Energy contribution | -18.96 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
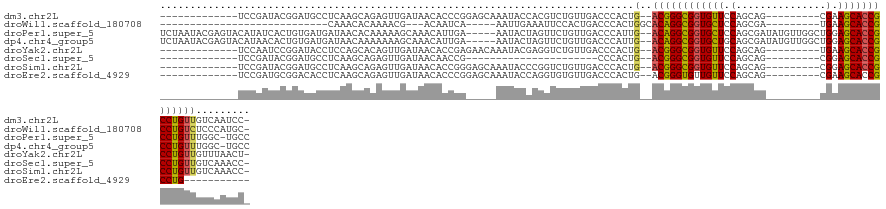
>dm3.chr2L 6351659 110 - 23011544 -------------UCCGAUACGGAUGCCUCAAGCAGAGUUGAUAACACCCGGAGCAAAUACCACGUCUGUUGACCCACUG--ACGGGCGGUGUUCCAGCAG---------CGAAGCACCGCCUGUUGUCAAUCC- -------------((((...))))(((.....)))..((((((((((...((........)).((((.((......)).)--)))(((((((((.(.....---------.).)))))))))))))))))))..- ( -35.50, z-score = -1.57, R) >droWil1.scaffold_180708 5398044 89 + 12563649 ----------------------------CAAACACAAAACG---ACAAUCA-----AAUUGAAAUUCCACUGACCCACUGGCACAGGCGGUGCUCCAGCGA---------UGAAGCACCGCCUGUCUCCCAUGC- ----------------------------.............---.......-----......................(((.((((((((((((.((....---------)).))))))))))))...)))...- ( -27.30, z-score = -3.43, R) >droPer1.super_5 367517 127 + 6813705 UCUAAUACGAGUACAUAUCACUGUGAUGAUAACACAAAAAGCAAACAUUGA-----AAUACUAGUUCUGUUGACCCAUUG--ACAGGCGGUGCUCCAGCGAUAUGUUGGCUGGAGCACCGCCUGUUUGGC-UGCC ...............(((((......))))).........(((((((..((-----(.......)))))))...(((..(--(((((((((((((((((.(.....).))))))))))))))))))))).-))). ( -46.60, z-score = -4.53, R) >dp4.chr4_group5 371564 127 + 2436548 UCUAAUACGAGUACAUAACACUGUGAUGAUAACAAAAAAAGCAAACAUUGA-----AAUACUAGUUCUGUUGACCCAUUG--ACAGGCGGUGCUGCAGCGAUAUGUUGGCUGGAGCACCGCCUGUUUGGC-UGCC .............((((....))))...............(((((((..((-----(.......)))))))...(((..(--((((((((((((.((((.(.....).)))).)))))))))))))))).-))). ( -40.70, z-score = -2.95, R) >droYak2.chr2L 15771633 110 + 22324452 -------------UCCAAUCCGGAUACCUCCAGCACAGUUGAUAACACCGAGAACAAAUACGAGGUCUGUUGACCCACUG--ACGGGCGGUGUUCCAGCAG---------UGAAGCACCGCCUGUUGUUUAACU- -------------........(((....))).....((((((...................(.((((....)))))...(--((((((((((((.((....---------)).)))))))))))))..))))))- ( -34.00, z-score = -1.85, R) >droSec1.super_5 4422289 88 - 5866729 -------------UCCGAUACGGAUGCCUCAAGCAGAGUUGAUAACAACCG----------------------CCCACUG--ACGGGCGGUGUUCCAGCAG---------CGGAGCACCGCCUGUUGUCAAACC- -------------...(((..((.(((.....)))(.((((....))))).----------------------.))...(--((((((((((((((.....---------.)))))))))))))))))).....- ( -36.30, z-score = -3.50, R) >droSim1.chr2L 6158513 110 - 22036055 -------------UCCGAUACGGAUGCCUCAAGCAGAGUUGAUAACACCGGGAGCAAAUACCCGGUCUGUUGACCCACUG--ACGGGCGGUGUUCCAGCAG---------CGGAGCACCGCCUGUUGUCAAACC- -------------...(((..((.(((.....)))..((..(((..((((((........)))))).)))..))))...(--((((((((((((((.....---------.)))))))))))))))))).....- ( -49.00, z-score = -4.31, R) >droEre2.scaffold_4929 15280183 100 - 26641161 -------------UCCGAUGCGGACACCUCAAGCAGAGUUGAUAACACCCGGAGCAAAUACCAGGUGUGUUGACCCACUG--ACGGGUGUUGUUCCAGCAG---------CGAAGCACCGCCUG----------- -------------......((((((((((....(((.((..(((.((((.((........)).)))))))..))...)))--..)))))).......((..---------....)).))))...----------- ( -31.00, z-score = -0.86, R) >consensus _____________UCCGAUACGGAUACCUCAAGCAGAGUUGAUAACACCCA_____AAUACCAGGUCUGUUGACCCACUG__ACGGGCGGUGUUCCAGCAG_________CGAAGCACCGCCUGUUGUCAAUCC_ ..................................................................................((((((((((((.(...............).)))))))))))).......... (-19.29 = -18.96 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:06 2011