| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,132,706 – 1,132,847 |
| Length | 141 |
| Max. P | 0.724568 |

| Location | 1,132,706 – 1,132,847 |
|---|---|
| Length | 141 |
| Sequences | 7 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 68.93 |
| Shannon entropy | 0.56483 |
| G+C content | 0.41114 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -13.31 |
| Energy contribution | -9.90 |
| Covariance contribution | -3.41 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
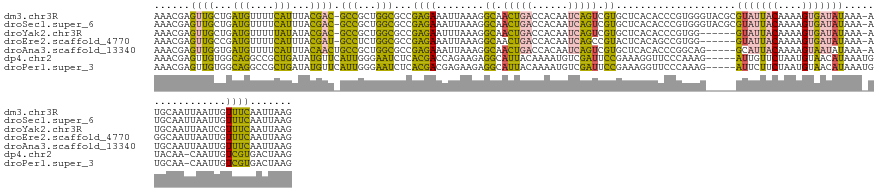
>dm3.chr3R 1132706 141 - 27905053 AAACGAGUUGCUGAUGUUUUCAUUUACGAC-GCCGCUGGCGCCGAGAAAUUAAAGGCAACUGACCACAAUCAGUCGUGCUCACACCCGUGGGUACGCGUAUUACAAAAGUGAUAUAAA-AUGCAAUUAAUUGUUUCAAUUAAG ......((((..((((....))))..))))-(((...)))...((((((((((..((((((((......)))))(((((((((....))))))))).(((((((....)))))))...-.)))..)))))).))))....... ( -38.30, z-score = -1.87, R) >droSec1.super_6 1254145 141 - 4358794 AAACGAGUUGCUGAUGUUUUCAUUUACGAC-GCCGCUGGCGCCGAGAAAUUAAAGGCAACUGACCACAAUCAGUCGUGCUCACACCCGUGGGUACGCGUAUUACAAAAGUGAUAUAAA-AUGCAAUUAAUUGUUUCAAUUAAG ......((((..((((....))))..))))-(((...)))...((((((((((..((((((((......)))))(((((((((....))))))))).(((((((....)))))))...-.)))..)))))).))))....... ( -38.30, z-score = -1.87, R) >droYak2.chr3R 1513900 135 - 28832112 AAACGAGUUGCUGAUGUUUUUAUAUACGAC-GCCGCUGGCGCCGAGAAUUUAAAGGCAACUGACCACAAUCAGUCGUGCUCACACCCGUGG------GUAUUACAAAAGUGAUAUAAA-AUGCAAUUAAUCGUUUCAAUUAAG (((((((((((......(((((((((((((-(((...)))(((...........)))...............)))))((((((....))))------))............)))))))-).)))))...))))))........ ( -32.40, z-score = -1.06, R) >droEre2.scaffold_4770 1407566 135 - 17746568 AAACGAGUUGCCGAUGUUUUCAUUUACGAU-GCCUCUGGCGCCGAGAAAUUAAAGGCAACUGACCACAAUCAGCCGUACUCACAGCCGUGG------GUAUUACAAAAGUGAUAUAAA-AGGCAAUUAAUUGUUUCAAUUAAG ((((((((((((.......((((((..(((-((((.(((((((...........)))..((((......))))...........)))).))------)))))....))))))......-.))))))...))))))........ ( -35.94, z-score = -2.33, R) >droAna3.scaffold_13340 23314527 137 - 23697760 AAACGAGUUGGUGAUGUUUUCAUUUACAACUGCCGCUGGCGCCGAGAAAUUAAAGGCAACUGACCACAAUCAGUCGUGCUCACACCCGGCAG-----GCAUUACAAAAGUAAUAUAAA-AUGCAAUUAAUUGUUUCAAUUAAG .....(((((..((((....))))..)))))((((((((.(((...........))).(((((......))))).(((....)))))))).)-----))(((((....))))).....-......(((((((...))))))). ( -32.70, z-score = -1.17, R) >dp4.chr2 9141310 137 + 30794189 AAACGAGUUGUGGCAGGCCGCUGAUAUGUUCAUUGGGAAUCUCACGACCAGAAGAGGCAUUACAAAAUGUCGAUUCCGAAAGGUUCCCAAAG-----AUUGUUCUAAUGUAACAUAAAUGUACAA-CAAUUGUCGUGACUAAG .....(((..((((((...((...((((((((((((((((((..((....(((..((((((....))))))..)))))..))))))))..((-----(....))))))).))))))...))....-...))))))..)))... ( -38.00, z-score = -2.24, R) >droPer1.super_3 2938286 137 + 7375914 AAACGAGUUGUGGCAGGCCGCUGAUAUGUUCAUUGGGAAUCUCACGACGAGAAGAGGCAUUACAAAAUGUCGAUUCCGAAAGGUUCCCAAAG-----AUUCUUCUAAUGUAACAUAAAUGUGCAA-CAAUUGUCGUGACUAAG .....(((..((((((..(((...((((((((((((((((((.....((.(((..((((((....))))))..)))))..))))))))..((-----(....))))))).))))))...)))...-...))))))..)))... ( -40.70, z-score = -2.77, R) >consensus AAACGAGUUGCUGAUGUUUUCAUUUACGAC_GCCGCUGGCGCCGAGAAAUUAAAGGCAACUGACCACAAUCAGUCGUGCUCACACCCGUGGG_____GUAUUACAAAAGUGAUAUAAA_AUGCAAUUAAUUGUUUCAAUUAAG ......((((...(((....)))...)))).(((...)))...((((........((.(((((......))))).))....................(((((((....))))))).................))))....... (-13.31 = -9.90 + -3.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:13 2011