
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,108,646 – 1,108,745 |
| Length | 99 |
| Max. P | 0.792678 |

| Location | 1,108,646 – 1,108,745 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 63.66 |
| Shannon entropy | 0.70441 |
| G+C content | 0.52109 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -9.09 |
| Energy contribution | -8.87 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1108646 99 - 27905053 AGAGAGAGGCGAUA-UGUGGAGCGUUAGAGCUUUCAA---ACAGUGCUAGCCCA-AAAAGUGGGACACCCCCACG-CACGAAAGCUUUUG-CAUCUGCUACCUCUG ....(((((.....-.(..((..((.(((((((((..---...((((.......-....(((((.....))))))-)))))))))))).)-).))..)..))))). ( -32.30, z-score = -2.10, R) >droSim1.chr3R 1132360 91 - 27517382 --------GCGAUA-UGUGGAGCGCCAGAGCUUUCGA---ACAGUGCUAGCCCA-AAUAGUGGGACACCCCCACG-CACGAAAGCUUUUG-CAGCUGCUACCGCUG --------(((...-.(((((((((.((((((((((.---...((....))...-....(((((.....))))).-..)))))))))).)-).))).))))))).. ( -34.20, z-score = -2.98, R) >droSec1.super_6 1229909 91 - 4358794 --------GCGAUA-UGUGGAGCGCCAGAGCUUUCGA---ACAGUGCUAGCCCA-AAUAGUGGGACACCCCCACG-CACGAAAGCUUUUG-CAGCUGCUACCGCUG --------(((...-.(((((((((.((((((((((.---...((....))...-....(((((.....))))).-..)))))))))).)-).))).))))))).. ( -34.20, z-score = -2.98, R) >droYak2.chr3R 1488873 87 - 28832112 --------GCGAUA-UGUGGAGCGCCAAAGCUUUCUC---ACAGUGCUAGCCCA-AAAAGUGGGACAUCCCCAC---ACGAAAGCUUUUG-CAGCUGCUACCGC-- --------(((...-.(((((((((.(((((((((..---...((....))...-....(((((.....)))))---..))))))))).)-).))).)))))))-- ( -30.40, z-score = -2.47, R) >droEre2.scaffold_4770 1383814 90 - 17746568 --------GCGAUA-UGUGGAGCGCCAGAGCUUUCGC---ACAGUGCUAGCCCA-AAAAGUGGGACA-CCCCACA-CACGAAAGCUUUUG-CAGCUGCUACCGCUG --------(((...-.(((((((((.(((((((((((---.....)).......-....(((((...-.))))).-...))))))))).)-).))).))))))).. ( -34.60, z-score = -2.94, R) >droAna3.scaffold_13340 23291808 86 - 23697760 --------GCGAGAGCAAUGAGCGCCAAAGCUCCAAC---ACAGUGCUAGCCAU-AAAAGUGGGACACCCUC--------AAAGCUUUGGUCAGCUGUUUCUUGUU --------(((((((((..((((......))))....---.....(((..(((.-.....((((.....)))--------)......)))..))))).))))))). ( -24.40, z-score = -0.61, R) >dp4.chr2 9117142 86 + 30794189 --------ACAAUGCUGCUGAGCGGCAAAGCUCUCACACUACACUGCUAGCCAA-AAAAGUUGGACAAGCU---------UGAGCUUUUGCC--UAACUACCGUUG --------.(((((((....)))((((((((((............(((.(((((-.....)))).).))).---------.)))).))))))--........)))) ( -21.94, z-score = -0.52, R) >droPer1.super_3 2914075 86 + 7375914 --------ACAAUGCUGCUGAGCGGGAAAGCUCUCACACUACACUGCUAGCCAA-AAAAGUUGGACAGGCU---------UGAGCUUUUGCC--UAACUACCGUUG --------.(((((......((.(.((((((((..........((((((((...-....))))).)))...---------.)))))))).))--)......))))) ( -20.74, z-score = 0.18, R) >droWil1.scaffold_181130 3839759 87 + 16660200 ---------------GCAAAAGCUCCACCACCCACAAAUGCUGCCGCUAAAUGGCUAAAGCUCUACACUAACAGGCCAUAAAAGUGUGCAUAGUUUGCUCUU---- ---------------((((((((................)))((((((..((((((.................))))))...)))).))....)))))....---- ( -17.72, z-score = -0.01, R) >consensus ________GCGAUA_UGUGGAGCGCCAAAGCUUUCAC___ACAGUGCUAGCCCA_AAAAGUGGGACACCCCCAC__CACGAAAGCUUUUG_CAGCUGCUACCGCUG ................(((((((...(((((((..........(((....((((......)))).))).............)))))))........))).)))).. ( -9.09 = -8.87 + -0.23)

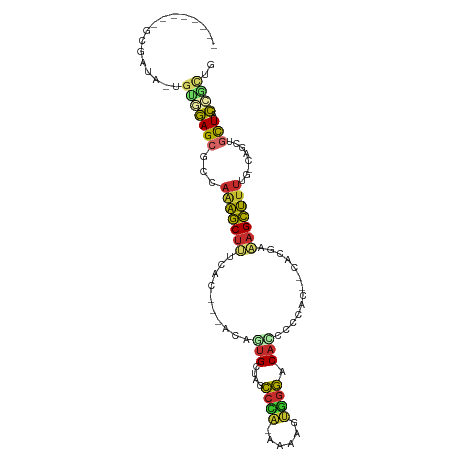
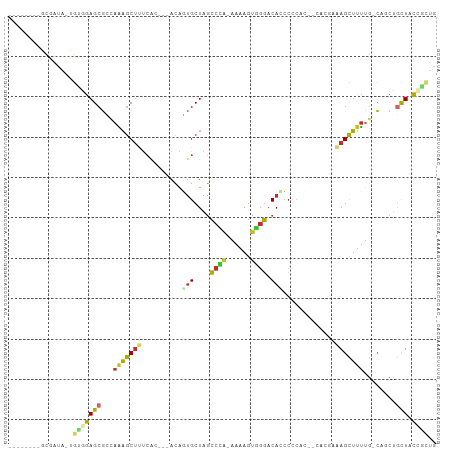
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:06 2011