| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,090,099 – 1,090,253 |
| Length | 154 |
| Max. P | 0.933497 |

| Location | 1,090,099 – 1,090,253 |
|---|---|
| Length | 154 |
| Sequences | 5 |
| Columns | 163 |
| Reading direction | forward |
| Mean pairwise identity | 79.79 |
| Shannon entropy | 0.36061 |
| G+C content | 0.44487 |
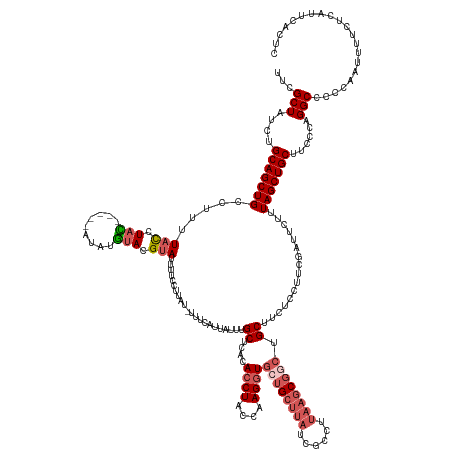
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -19.54 |
| Energy contribution | -21.34 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
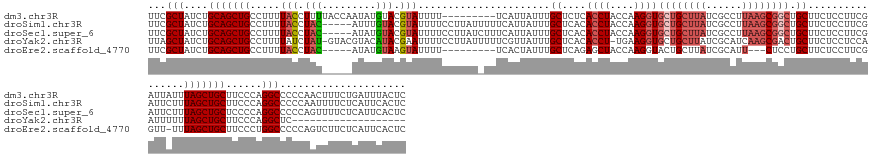
>dm3.chr3R 1090099 154 + 27905053 UUCGCUAUCUGCAGCUGCCUUUUACCUUUUACCAAUAUGUACGUAUUUU---------UCAUUAUUUGCUCUCACCUACCAAGGUGCUGCUUAUCGCCUUAAGCGGCUGCUUCUCCUUCGAUUAUUUAGCUGCUUCCCAGGCCCCCAACUUUCUGAUUUACUC ...(((....(((((((......(((((..........(((.(((....---------....))).)))...........)))))((((((((......))))))))...................)))))))......)))..................... ( -29.40, z-score = -1.65, R) >droSim1.chr3R 1114124 158 + 27517382 UUCGCUAUCUGCAGCUGCCUUUUACCUAC-----AUUUGUACGUAUUUUCCUUAUUUUUCAUUAUUUGCUCACACCUACCAAGGUGCUGCUUAUCGCCUUAAGCGGCUGCUUCUCCUUCGAUUCUUUAGCUGCUUCCCAGGCCCCCAAUUUUCUCAUUCACUC ...(((....(((((((.....(((.(((-----....))).)))......................((....((((....))))((((((((......)))))))).))................)))))))......)))..................... ( -30.90, z-score = -2.96, R) >droSec1.super_6 1211439 158 + 4358794 UUCGCUAUCUGCAGCUGCCUUUUACCUAC-----AUAUGUACGUAUUUUCCUUAUCUUUCAUUAUUUGCUCACACCUACCAAGGUGCUGCUUAUCGCCUUAAGCGGCUGCUUCUCCUUCGAUUCUUUAGCUGCUCCCCAGGCCCCCAGUUUUCUCAUUCACUC ...(((....(((((((.....(((.(((-----....))).)))......................((....((((....))))((((((((......)))))))).))................)))))))......)))..................... ( -30.90, z-score = -2.39, R) >droYak2.chr3R 1462412 142 + 28832112 UUAGCUAUCUGCAGCUGCCUUUUAUCUAU-GUACGUACAUACGAAUUUUCCUUAUUUUUCGUUAUUUGCUCACACCU-UGAAGGUGCUGCUUAUCGCAUCAAGCGACUGCUUCUCCUCCAAUUUUUUAGCUGCUUCCCAGGCUC------------------- ..(((((...(((((.((((((......(-((..(((.(((((((............)))).))).)))..)))...-.)))))))))))...((((.....))))....................)))))(((.....)))..------------------- ( -31.70, z-score = -2.36, R) >droEre2.scaffold_4770 1365561 145 + 17746568 UUCGCUAUCUGCAGCUGCCUUUUACCUAC-----AUAUGUAAGUAUUUU---------UCACUAUUUGCUCAGAGCUACCAAGGUACUGCUUAUCGCAUU---CUCCUGCUUCUCCUUCGGUU-UUUAGCUGCUUCCCUGGCCCCCAGUCUUCUCAUUCACUC ...((((...(((((((......(((...-----....(((((((....---------....)))))))...(((......(((...(((.....)))..---..)))....)))....))).-..))))))).....))))..................... ( -26.20, z-score = -0.97, R) >consensus UUCGCUAUCUGCAGCUGCCUUUUACCUAC_____AUAUGUACGUAUUUUCCUUAU_UUUCAUUAUUUGCUCACACCUACCAAGGUGCUGCUUAUCGCCUUAAGCGGCUGCUUCUCCUUCGAUUCUUUAGCUGCUUCCCAGGCCCCCAAUUUUCUCAUUCACUC ...(((....(((((((........................................................((((....))))((((((((......))))))))...................)))))))......)))..................... (-19.54 = -21.34 + 1.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:51:03 2011