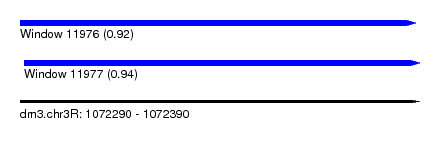
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,072,290 – 1,072,390 |
| Length | 100 |
| Max. P | 0.943425 |

| Location | 1,072,290 – 1,072,389 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Shannon entropy | 0.13378 |
| G+C content | 0.47554 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -22.28 |
| Energy contribution | -21.80 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
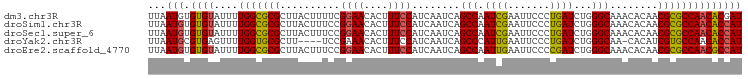
>dm3.chr3R 1072290 99 + 27905053 CUUAAUGUGUGUAUUUUGGCGCGCUUACUUUUCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACGA .......(((((....(((((((..........((((....))))........(((.((((.......))))...)))........)))))))))))). ( -26.10, z-score = -2.42, R) >droSim1.chr3R 1098624 99 + 27517382 CUUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACCA .....((.((((....(((((((..........((((....))))........(((.((((.......))))...)))........))))))))))))) ( -25.60, z-score = -2.51, R) >droSec1.super_6 1196063 99 + 4358794 CUUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACCA .....((.((((....(((((((..........((((....))))........(((.((((.......))))...)))........))))))))))))) ( -25.60, z-score = -2.51, R) >droYak2.chr3R 1439893 94 + 28832112 CUUAAUGCGUGAGUUUUGGUGCGCUU----UCCGAAACACUUUCCAUCAAUCAGCCCAUUGAAUUCCCUGAUCUGGGCAA-CACAUCGUGCCAACACCA ........(((.(((((((.......----.)))))))...............(((((...............)))))..-)))...(((....))).. ( -19.36, z-score = -0.21, R) >droEre2.scaffold_4770 1349338 99 + 17746568 CUUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUUGAAUUCCCCGAUCUGGGCAAACACAACGCGCCAACGCCA .....((.((((....(((((((..........((((....))))........(((.((((.......))))...)))........))))))))))))) ( -25.10, z-score = -1.67, R) >consensus CUUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACCA ........((((....(((((((..........((((....))))........(((.((((.......))))...)))........))))))))))).. (-22.28 = -21.80 + -0.48)

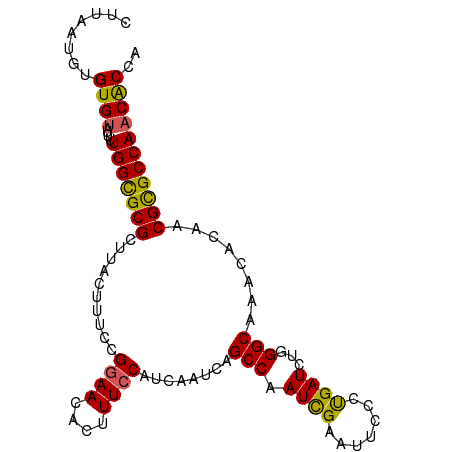
| Location | 1,072,291 – 1,072,390 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Shannon entropy | 0.13378 |
| G+C content | 0.46533 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1072291 99 + 27905053 UUAAUGUGUGUAUUUUGGCGCGCUUACUUUUCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACGAU ......(((((....(((((((..........((((....))))........(((.((((.......))))...)))........)))))))))))).. ( -26.10, z-score = -2.38, R) >droSim1.chr3R 1098625 99 + 27517382 UUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACCAU ...(((.((((....(((((((..........((((....))))........(((.((((.......))))...)))........)))))))))))))) ( -26.70, z-score = -2.90, R) >droSec1.super_6 1196064 99 + 4358794 UUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACCAU ...(((.((((....(((((((..........((((....))))........(((.((((.......))))...)))........)))))))))))))) ( -26.70, z-score = -2.90, R) >droYak2.chr3R 1439894 94 + 28832112 UUAAUGCGUGAGUUUUGGUGCGCUU----UCCGAAACACUUUCCAUCAAUCAGCCCAUUGAAUUCCCUGAUCUGGGCAA-CACAUCGUGCCAACACCAU .......(((.(((((((.......----.)))))))...............(((((...............)))))..-)))...(((....)))... ( -19.36, z-score = -0.24, R) >droEre2.scaffold_4770 1349339 99 + 17746568 UUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUUGAAUUCCCCGAUCUGGGCAAACACAACGCGCCAACGCCAU ...(((.((((....(((((((..........((((....))))........(((.((((.......))))...)))........)))))))))))))) ( -26.20, z-score = -2.05, R) >consensus UUAAUGUGUGUAUUUUGGCGCGCUUACUUUCCGGAACACUUUCCAUCAAUCAGCCAAUCGAAUUCCCUGAUCUGGGCAAACACAACGCGCCAACACCAU ...(((.((((....(((((((..........((((....))))........(((.((((.......))))...)))........)))))))))))))) (-22.46 = -22.18 + -0.28)
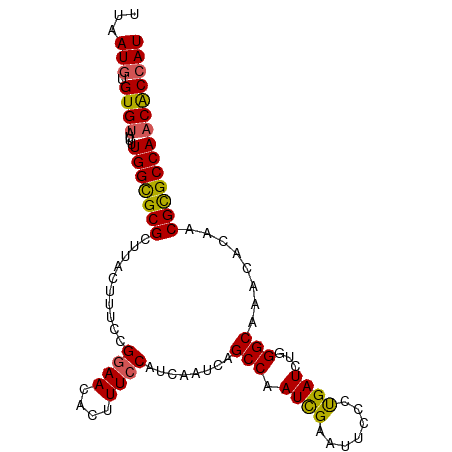
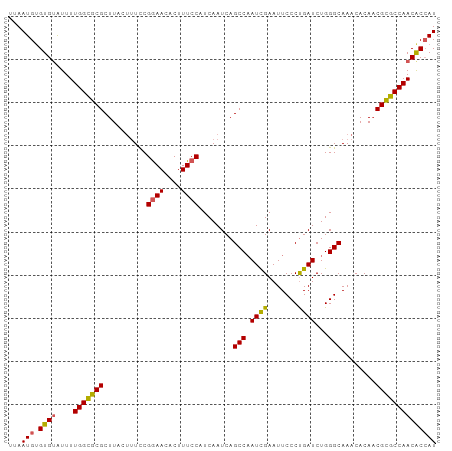
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:57 2011