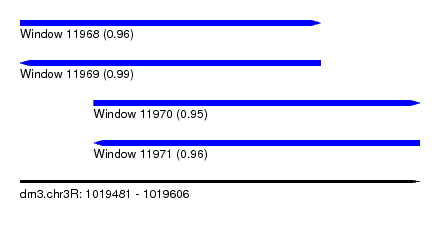
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 1,019,481 – 1,019,606 |
| Length | 125 |
| Max. P | 0.994972 |

| Location | 1,019,481 – 1,019,575 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.33 |
| Shannon entropy | 0.52931 |
| G+C content | 0.50218 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -16.70 |
| Energy contribution | -18.28 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
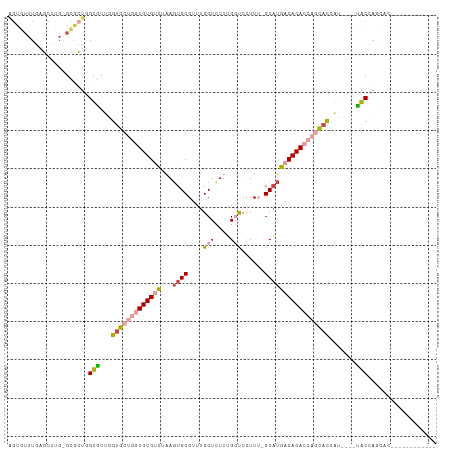
>dm3.chr3R 1019481 94 + 27905053 AGUGUUUGAGCUUG-GCGCUAGUGUUGGUGCUGGUGUGUCUGAGUGCGUUUGCUCUUUGGUCCUUU-GCAUGACACACCAGCACCAU----UACCACCAC------------ ((((((.......)-))))).(((.(((((((((((((((...(((((...(((....)))....)-))))))))))))))))))).----)))......------------ ( -40.60, z-score = -3.52, R) >droSec1.super_6 1142485 94 + 4358794 AGUGUUUGAGCUUG-GCGCUAGUGUUGGUGCUGGUGUGUCUAAGUGCGUUUGCUCUUUGGUCCUUU-GCAUGACACACCAGCACCAU----UGCCAGCAC------------ .........(((.(-((((....))(((((((((((((((...(((((...(((....)))....)-))))))))))))))))))).----.))))))..------------ ( -42.60, z-score = -3.68, R) >droYak2.chr3R 1379632 93 + 28832112 AGUGUUUGAGCUCG-GCGCUGGUAUUAGUGCUGGUGUGUCUAACUGCGUUUGCUCUUUGGUCCUU--GCAUGACACACCAGCACCAU----UACCAUUAC------------ ((((((.......)-)))))((((...(((((((((((((....((((...(((....)))...)--))).)))))))))))))...----)))).....------------ ( -38.20, z-score = -4.03, R) >droEre2.scaffold_4770 1294812 109 + 17746568 AGUGUUUGAGCUCG-GCACUGGUGUUGGUGCUGGUGUGUCUAAGUGCGUUUGCUCUUUGGUCCUU--GCAUGACACACCAGCACAUCAGCACACCAGCACCAUUACCAACAC .(((((.......(-(..(((((((..(((((((((((((...(((((...(((....)))...)--)))))))))))))))))......)))))))..))......))))) ( -45.32, z-score = -2.74, R) >droAna3.scaffold_13340 23214166 85 + 23697760 GGUGUUUGCUGAAGGGUGAUGGUGUUGGUG----UGUGCUCAAGUGCGUUUGCUUUUGGUCCUUUUUGCCUGACACACUGCCAU-AU----UAC------------------ .(((((.((.((((((.(((.(....((((----..(((......)))..))))..).)))))))))))..)))))........-..----...------------------ ( -19.10, z-score = 1.04, R) >dp4.chr2 9022104 85 - 30794189 ------UGUGUGUGUGUGCUGGUAUUGGUG----UGUGCUUAAGUGCGUUUGCUUUUGG-CCCUUUUGCAUGACACACAACUUUUGC----UGCCAGAGA------------ ------..(((((((((((.(((...((((----..(((......)))..))))....)-)).....)))).)))))))..(((((.----...))))).------------ ( -22.40, z-score = 0.45, R) >consensus AGUGUUUGAGCUUG_GCGCUGGUGUUGGUGCUGGUGUGUCUAAGUGCGUUUGCUCUUUGGUCCUUU_GCAUGACACACCAGCACCAU____UACCAGCAC____________ ...........................(((((((((((((...((((....................)))))))))))))))))............................ (-16.70 = -18.28 + 1.58)


| Location | 1,019,481 – 1,019,575 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.33 |
| Shannon entropy | 0.52931 |
| G+C content | 0.50218 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -15.28 |
| Energy contribution | -17.33 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1019481 94 - 27905053 ------------GUGGUGGUA----AUGGUGCUGGUGUGUCAUGC-AAAGGACCAAAGAGCAAACGCACUCAGACACACCAGCACCAACACUAGCGC-CAAGCUCAAACACU ------------(((.((((.----.(((((((((((((((.((.-.......))..(((........))).)))))))))))))))..)))).)))-.............. ( -39.00, z-score = -4.04, R) >droSec1.super_6 1142485 94 - 4358794 ------------GUGCUGGCA----AUGGUGCUGGUGUGUCAUGC-AAAGGACCAAAGAGCAAACGCACUUAGACACACCAGCACCAACACUAGCGC-CAAGCUCAAACACU ------------(((((((..----.(((((((((((((((.((.-.......))(((.((....)).))).)))))))))))))))...)))))))-.............. ( -41.90, z-score = -5.05, R) >droYak2.chr3R 1379632 93 - 28832112 ------------GUAAUGGUA----AUGGUGCUGGUGUGUCAUGC--AAGGACCAAAGAGCAAACGCAGUUAGACACACCAGCACUAAUACCAGCGC-CGAGCUCAAACACU ------------....(((((----.(((((((((((((((.(((--..................)))....))))))))))))))).)))))((..-...))......... ( -37.67, z-score = -4.66, R) >droEre2.scaffold_4770 1294812 109 - 17746568 GUGUUGGUAAUGGUGCUGGUGUGCUGAUGUGCUGGUGUGUCAUGC--AAGGACCAAAGAGCAAACGCACUUAGACACACCAGCACCAACACCAGUGC-CGAGCUCAAACACU ((((((((..(((..(((((((......(((((((((((((.((.--......))(((.((....)).))).)))))))))))))..)))))))..)-)).)))..))))). ( -50.80, z-score = -4.17, R) >droAna3.scaffold_13340 23214166 85 - 23697760 ------------------GUA----AU-AUGGCAGUGUGUCAGGCAAAAAGGACCAAAAGCAAACGCACUUGAGCACA----CACCAACACCAUCACCCUUCAGCAAACACC ------------------...----..-.(((..(((((((((((......).))....((....))...))).))))----).)))......................... ( -13.30, z-score = 0.50, R) >dp4.chr2 9022104 85 + 30794189 ------------UCUCUGGCA----GCAAAAGUUGUGUGUCAUGCAAAAGGG-CCAAAAGCAAACGCACUUAAGCACA----CACCAAUACCAGCACACACACACA------ ------------.........----......((.((((((..(((......(-(.....))....((......))...----...........))).)))))))).------ ( -14.30, z-score = 1.13, R) >consensus ____________GUGCUGGUA____AUGGUGCUGGUGUGUCAUGC_AAAGGACCAAAGAGCAAACGCACUUAGACACACCAGCACCAACACCAGCGC_CAAGCUCAAACACU ............................(((((((((((((.((.........))....((....)).....)))))))))))))........................... (-15.28 = -17.33 + 2.06)
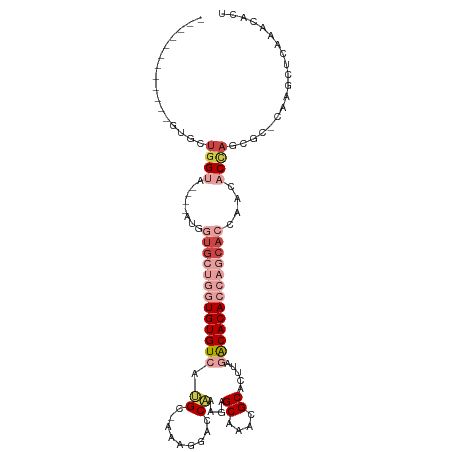
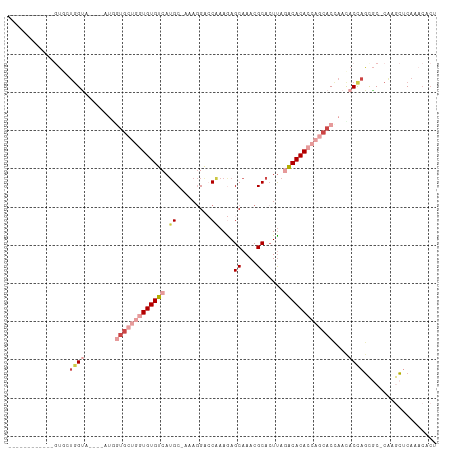
| Location | 1,019,504 – 1,019,606 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.99 |
| Shannon entropy | 0.58747 |
| G+C content | 0.51692 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -15.38 |
| Energy contribution | -17.18 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1019504 102 + 27905053 UUGGUGCUGGUGUGUCUGAGUGCGUUUGCUCUUUGGUCCUUU--GCAUGACACACCAGCAC----------------CAUUACCACCACGCCACCAGCGGCAGUGGCAGUGGCAGUAGCA .(((((((((((((((...(((((...(((....)))....)--)))))))))))))))))----------------))((((..(((((((((........))))).))))..)))).. ( -50.30, z-score = -3.63, R) >droYak2.chr3R 1379655 89 + 28832112 UUAGUGCUGGUGUGUCUAACUGCGUUUGCUCUUUGGUCCUU---GCAUGACACACCAGCAC----------------CAUUACCAUUACGCCCCCAGUGGCAUUGGCA------------ ...(((((((((((((....((((...(((....)))...)---))).)))))))))))))----------------.....(((....(((......)))..)))..------------ ( -34.70, z-score = -3.33, R) >droEre2.scaffold_4770 1294835 105 + 17746568 UUGGUGCUGGUGUGUCUAAGUGCGUUUGCUCUUUGGUCCUU---GCAUGACACACCAGCACAUCAGCACACCAGCACCAUUACCAACACGCCACCAGUGGCAGUGCCA------------ .((((((((((((((....((((...(((............---)))((......))))))....)))))))))))))).......(((((((....)))).)))...------------ ( -42.40, z-score = -2.63, R) >droAna3.scaffold_13340 23214190 92 + 23697760 UUGGUG----UGUGCUCAAGUGCGUUUGCU-UUUGGUCCUUUUUGCCUGACACACUGCCAU-----------------AUUACAGUAGCAGUGGCAGUGGCAGUUUCAGGAUAU------ ..((((----..(((......)))..))))-....(((((..(((((.....((((((.((-----------------......)).)))))).....)))))....)))))..------ ( -26.50, z-score = 0.06, R) >dp4.chr2 9022122 90 - 30794189 UUGGUG----UGUGCUUAAGUGCGUUUGCU-UUUGGCCCUUUU-GCAUGACACACAACUUU----------------UGCUGCCAGAGAGCGAGAGAGAGAGAGGACACUCA-------- ((((((----(((((..(((.((.......-....)).)))..-)))).)))).)))((((----------------((((.......)))))))).....(((....))).-------- ( -25.50, z-score = -0.08, R) >consensus UUGGUGCUGGUGUGUCUAAGUGCGUUUGCUCUUUGGUCCUUU__GCAUGACACACCAGCAC________________CAUUACCAUAACGCCACCAGUGGCAGUGGCA____________ ...(((((((((((((...((((....(((....))).......)))))))))))))))))........................................................... (-15.38 = -17.18 + 1.80)

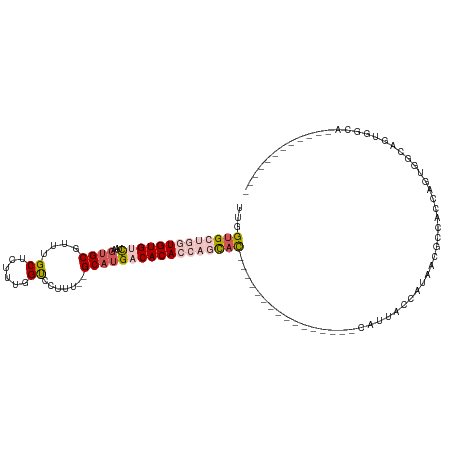
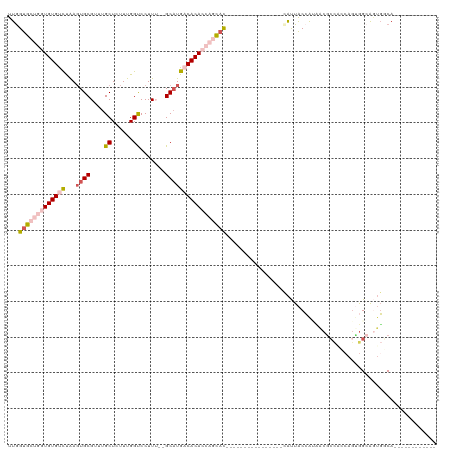
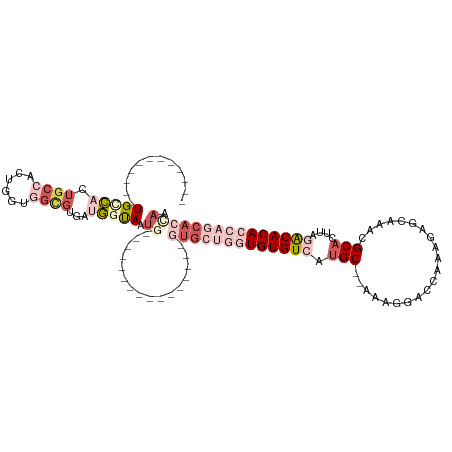
| Location | 1,019,504 – 1,019,606 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.99 |
| Shannon entropy | 0.58747 |
| G+C content | 0.51692 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -13.48 |
| Energy contribution | -17.16 |
| Covariance contribution | 3.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 1019504 102 - 27905053 UGCUACUGCCACUGCCACUGCCGCUGGUGGCGUGGUGGUAAUG----------------GUGCUGGUGUGUCAUGC--AAAGGACCAAAGAGCAAACGCACUCAGACACACCAGCACCAA ...(((..((((.((((((......))))))))))..))).((----------------(((((((((((((.((.--.......))..(((........))).))))))))))))))). ( -54.30, z-score = -4.89, R) >droYak2.chr3R 1379655 89 - 28832112 ------------UGCCAAUGCCACUGGGGGCGUAAUGGUAAUG----------------GUGCUGGUGUGUCAUGC---AAGGACCAAAGAGCAAACGCAGUUAGACACACCAGCACUAA ------------((((((((((......)))))..))))).((----------------(((((((((((((.(((---..................)))....))))))))))))))). ( -39.97, z-score = -4.03, R) >droEre2.scaffold_4770 1294835 105 - 17746568 ------------UGGCACUGCCACUGGUGGCGUGUUGGUAAUGGUGCUGGUGUGCUGAUGUGCUGGUGUGUCAUGC---AAGGACCAAAGAGCAAACGCACUUAGACACACCAGCACCAA ------------.(((((.((((....))))))))).....(((((((((((((((((.((((...((((((....---...)))......)))...)))))))).))))))))))))). ( -49.00, z-score = -2.93, R) >droAna3.scaffold_13340 23214190 92 - 23697760 ------AUAUCCUGAAACUGCCACUGCCACUGCUACUGUAAU-----------------AUGGCAGUGUGUCAGGCAAAAAGGACCAAA-AGCAAACGCACUUGAGCACA----CACCAA ------...((((....((((((((((((.(((....)))..-----------------.)))))))).).)))......)))).(((.-.((....))..)))......----...... ( -24.30, z-score = -1.23, R) >dp4.chr2 9022122 90 + 30794189 --------UGAGUGUCCUCUCUCUCUCUCGCUCUCUGGCAGCA----------------AAAGUUGUGUGUCAUGC-AAAAGGGCCAAA-AGCAAACGCACUUAAGCACA----CACCAA --------.(((((..............)))))....(((((.----------------...)))))((((..(((-..(((.((....-.......)).)))..))).)----)))... ( -15.84, z-score = 1.39, R) >consensus ____________UGCCACUGCCACUGGUGGCGUGAUGGUAAUG________________GUGCUGGUGUGUCAUGC__AAAGGACCAAAGAGCAAACGCACUUAGACACACCAGCACCAA ............(((((.((((......))))...)))))...................(((((((((((((...(......)........((....)).....)))))))))))))... (-13.48 = -17.16 + 3.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:52 2011