
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 968,483 – 968,580 |
| Length | 97 |
| Max. P | 0.821551 |

| Location | 968,483 – 968,580 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.17 |
| Shannon entropy | 0.46262 |
| G+C content | 0.54653 |
| Mean single sequence MFE | -36.09 |
| Consensus MFE | -20.45 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 968483 97 + 27905053 GGCCAUUAUCAGCGGCUGUCAAUAAGCGCAUUAAAA-CAUUUUUGCAAUUCGCGAGCUCGUUGACGGCUGAUGGCG-------------GAGACGCGCGGCUGUCAGGCAG- .(((.....(((((((((((((..((((........-)....((((.....)))))))..)))))))))....(((-------------....)))...))))...)))..- ( -37.80, z-score = -2.15, R) >droSim1.chr3R 992035 97 + 27517382 GGCCAUUAUCAGCGGCUGUCAAUAAGCGCAUUAAAA-CAUUUUUGCGAUUCGCGAGCUCGUUGACGGCUGAUGGCG-------------CAGACACGCGGCUGUCAGGCAG- .(((.........(((((((((..((((........-)....(((((...))))))))..)))))))))(((((((-------------(......)).)))))).)))..- ( -35.30, z-score = -1.47, R) >droSec1.super_6 1093001 97 + 4358794 GGCCAUUAUCAGCGGCUGUCAAUAAACGCAUUAAAA-CAUUUUUGCGAUUCGCGAGCUCGUUGACGGCUGAUGGCG-------------CAGACACGCGGCUGUCAGGCAG- .(((.........((((((((((...((((..((..-..))..))))...(....)...))))))))))(((((((-------------(......)).)))))).)))..- ( -32.80, z-score = -1.19, R) >droYak2.chr3R 1325261 73 + 28832112 GGCCAUUAGCAGCGGCUGUCAAUAAGCGCAUUAAAA-CAUUUUUGC------------------------AUGUCG-------------CAGACACGCGGCUGUCAGGCAG- .(((....(((((.((((((.....((((((((((.-....)))).------------------------))).))-------------).)))).)).)))))..)))..- ( -24.20, z-score = -1.82, R) >droEre2.scaffold_4770 1242825 110 + 17746568 GGCCAUUAUCGGCGGCUGUCAAUAAGCGCAUUAAAA-CAUUUUUGCAAUUCGCGAGCCCGUCGACGGCUGACGUCGACGGCUGACGGCGCAGACACGCGGCUGUCAGGCAG- .(((.....((((.((((((.....((((.......-......(((.....)))((((.(((((((.....)))))))))))....)))).)))).)).))))...)))..- ( -44.80, z-score = -2.13, R) >droAna3.scaffold_13340 23162610 96 + 23697760 CGCCAUUAUCGGCUGCUGUCAAUAAGCGCAUUAAAAACAUUUCUGCAAUUCGCGGGCUCGUUGACG---------------UGAUGGUGCUGACAAGCGGCUGUCGAACAG- ........(((((.(((((.....(((((((((..(((...(((((.....)))))...)))....---------------)))).))))).....))))).)))))....- ( -29.20, z-score = -0.55, R) >dp4.chr2 8963058 107 - 30794189 CGCCAUUAUCGGCGGCUGUCAAUAAGCGCAUUAAAAACAUUUCUGCAAUGCGAGAGCCUGUUGACGGUGGCGGUGA-----CGGUGGCGCUGACAUGCGGCUGUCAGGCAGA .(((.....(.((.((((((((((...(((..((.....))..)))...((....)).)))))))))).)).)(((-----((((.(((......))).))))))))))... ( -42.30, z-score = -1.57, R) >droPer1.super_3 2760990 107 - 7375914 CGCCAUUAUCGGCGGCUGUCAAUAAGCGCAUUAAAAACAUUUCUGCAAUGCGAGAGCCUGUUGACGGUGGCGGUGA-----CGGUGGCGCUGACAUGCGGCUGUCAGGCAGA .(((.....(.((.((((((((((...(((..((.....))..)))...((....)).)))))))))).)).)(((-----((((.(((......))).))))))))))... ( -42.30, z-score = -1.57, R) >consensus GGCCAUUAUCAGCGGCUGUCAAUAAGCGCAUUAAAA_CAUUUUUGCAAUUCGCGAGCUCGUUGACGGCUGAUGUCG_____________CAGACACGCGGCUGUCAGGCAG_ .(((.....((((.((((((.......(((..((.....))..)))...........(((....)))........................)))).)).))))...)))... (-20.45 = -20.26 + -0.19)

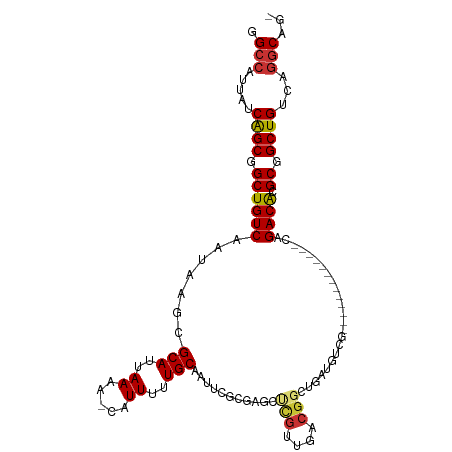
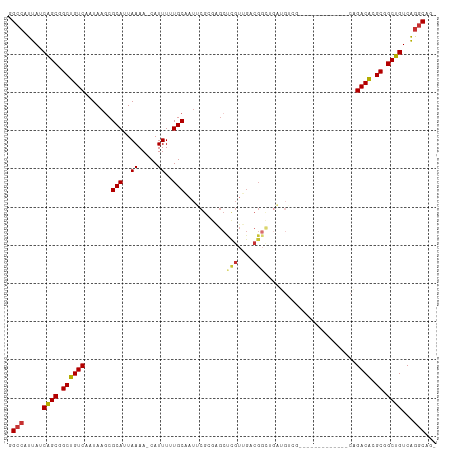
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:45 2011