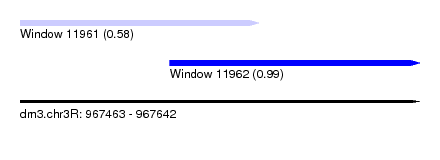
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 967,463 – 967,642 |
| Length | 179 |
| Max. P | 0.993337 |

| Location | 967,463 – 967,570 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Shannon entropy | 0.24030 |
| G+C content | 0.34918 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -16.42 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 967463 107 + 27905053 GCCCAGCGAAAGUUGUAUUGGGCUACAUCUUUUGUGGUUAAGUUAAUAUGUCAUUAGAGAGAAAUUUCAAAAUUAUAAUAUAGUAAGGAAGGGAGCACAAUUUAGAU (((((((....)).....)))))...((((.(((((.((...((............((((....))))....((((......)))).))...)).)))))...)))) ( -21.80, z-score = -1.52, R) >droSim1.chr3R 990951 93 + 27517382 ACCCAGCGAAAGUUGUAUUGGGCUACAUCAUUUGUAGUAAGGUUAAUAUGUCAUUAGAGAAAAGUUCAAAAA--------------AGAAGGGAGCACAAUUUAGAG .((((((....)))(((((((((((((.....))))))....))))))).......................--------------....))).............. ( -16.40, z-score = -1.03, R) >droSec1.super_6 1091918 93 + 4358794 ACCCAGCGAAAGUUGUAUUGGGCUACAUCAUUUGUAGUAAGGUUAAUAUGUCAUUAGAGAAAAGUUCAAAAA--------------AGGAGGGAGUACACUUUAGAG .((((((....)))(((((((((((((.....))))))....))))))).......................--------------....))).............. ( -16.40, z-score = -0.38, R) >consensus ACCCAGCGAAAGUUGUAUUGGGCUACAUCAUUUGUAGUAAGGUUAAUAUGUCAUUAGAGAAAAGUUCAAAAA______________AGAAGGGAGCACAAUUUAGAG .((((((....)))(((((((((((((.....))))))....))))))).........................................))).............. (-16.42 = -16.20 + -0.22)

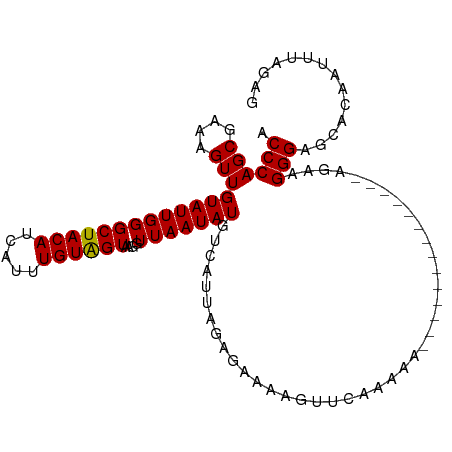
| Location | 967,530 – 967,642 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.71 |
| Shannon entropy | 0.27024 |
| G+C content | 0.34707 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -16.93 |
| Energy contribution | -18.93 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 967530 112 + 27905053 CAAAAUUAUAAUAUAGUAAGGAAGGGAGCACAAUUUAGAUCCCCUCUUUACAAGCCAUUAAUUUGUUUUUACCUAAC------AGCCCACAAUAUUCCAUUAACUGAAUAGUGCACCA ...............(((((((.((((.(........).)))).)))))))..(((......(((((.......)))------)).......(((((........)))))).)).... ( -18.60, z-score = -1.49, R) >droSim1.chr3R 991018 104 + 27517382 --------------AAAAAAGAAGGGAGCACAAUUUAGAGCCCCUCUUUACAAGCCAUUAAUUUGUUUUUACCUAAUUGUAAGAGCCCACACUAUUCAAUUUACUGAAUAGUGCACCA --------------...(((((.(((.((.(......).))))))))))...............((((((((......))))))))...(((((((((......)))))))))..... ( -26.10, z-score = -4.12, R) >droSec1.super_6 1091985 104 + 4358794 --------------AAAAAAGGAGGGAGUACACUUUAGAGCCCCUCUUUACAAGCCAUUAAUUUGUUUUUACCUAAUUGUAAAAGCACACACUAUUUAAUUUACUGAAUAGUGCGCCA --------------....((((((((.((.(......).))))))))))....((........(((((((((......)))))))))..(((((((((......))))))))).)).. ( -28.10, z-score = -3.88, R) >consensus ______________AAAAAAGAAGGGAGCACAAUUUAGAGCCCCUCUUUACAAGCCAUUAAUUUGUUUUUACCUAAUUGUAA_AGCCCACACUAUUCAAUUUACUGAAUAGUGCACCA .................((((.((((.((.(......).))))))))))...............((((((((......))))))))...(((((((((......)))))))))..... (-16.93 = -18.93 + 2.01)

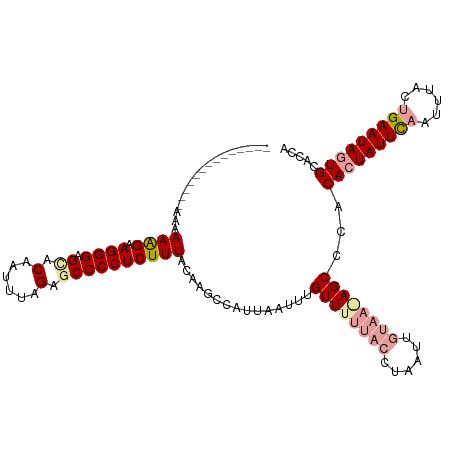
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:44 2011